Molecular Genetic Approach to the Fermented Horse Meat Microflora Screening
Klabukova DL1, Mashentseva NG2, Chernukha IМ2*, Fedulova LV2, Nikonov IN3, Laptev G.Yu3, Iljina LА3, Zamaratskaia G4, Jyldyrym ЕА3
1 FGBOU VPO “Moscow State University of Food Production”, Moscow, Russia.
2 V.M. Gorbatov Federal Research Center for Food Systems of Russian Academy of Sciences, Moscow, Russia.
3 LLC «BIOTROPH», Saint Petersburg, Russia.
4 Department of Food Science, Uppsala BioCenter, Swedish University of Agricultural Sciences, Uppsala, Sweden.
*Corresponding Author
Irina Chernukha,
V.M. Gorbatov Federal Research Center for Food Systems of Russian Academy of Sciences, Moscow, Russia.
E-mail: imcher@inbox.ru
Received: June 21, 2018; Accepted: July 19, 2018; Published: July 27, 2018
Citation: Klabukova DL, Mashentseva NG, Chernukha IМ, Fedulova LV, Nikonov IN, Laptev G.Yu, et al., Molecular Genetic Approach to the Fermented Horse Meat Microflora Screening. J Translational Diagn Technol. 2018;3(1):30-35.
Copyright: Chernukha IМ© 2017. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
The terminal restriction fragment length polymorphism (T-RFLP) technique is considered as a rapid and reliable tool for microbial community fingerprinting. The aim of the present study was to characterize microbial community of fermented sausage-like product from horse meat using T-RFLP technique. The product contained horse meat, horse fat, salt, honey and garlic. A total of 36 samples were collected, and 12 pooled samples were analysed. We demonstrated that microflora in the investigated fermented horse meat sausage-like product mainly consisted of safe and non-culturable bacteria (approximately 80%). The remaining part was represented by the conditionally pathogenic microflora, while the pathogenic microorganisms (Campylobacter) accounted only for .0.7% of total community. Lactobacteria accounted for 53.9% of the total microbial number when measured by T-RFLP, and 50.6% when using real-time PCR. We concluded that T-RFPL approaches can be effectively used for research purposes for detection of pathogenic and conditionally pathogenic microflora, and in inspection programs.
2.Abbreviations
3.Introduction
4.Materials and Methods
4.1 Fermented sausage-like product technology and sampling
4.2 DNA Extraction
4.3 PCR and T-RFLP
4.4 Real-time PCR
4.5 Statistical analysis
5.Results and Discussion
6.Conclusions
7.Financial Support
8.References
Keywords
T-RFLP; Horse Meat; Fermented Product; Microbial Community.
Abbreviations
T-RFLP: Terminal Restriction Fragment Length Polymorphism; PCR: Polymerase Chain Reaction; HRM: High Resolution Melting; LAB: Lactic Acid Bacteria; MAP: Modified Atmosphere Packaging.
Introduction
Nowadays, there is an increased awareness and demands among consumers for the safety of food products. Food quality control is one the major priorities for food industry and consumers to ensure high quality and safe production. Monitoring and assessment of microbiological quality is a primarily health-based activity to prevent the microbial spoilage and food poisoning, and protect public health.
Microbiological safety assessment of fermented meat products produced without thermal treatment requires special attention due to an increased risk for accumulation of pathogenic microflora. To prevent the microbial spoilage starter cultures with antibacterial properties are used in fermented sausage production.
Thus, it is also desirable to monitor starter cultures in order to control their development throughout the technological process and storage [1, 2]. Traditional microbiological and biochemical methods are usually cumbersome, time-consuming and often have limited accuracy. For example, traditional methods cannot detect non-culturable bacteria because of their low metabolism and resistance to the changes in environmental conditions. Nevertheless, non-culturable bacteria might remain viable and retain virulence. Therefore, information about the presence and diversity of the non-culturable forms of bacteria is important both for understanding of the ageing processes in fermented food and safety assessment [3].
To overcome issues of traditional methods, molecular genetic approaches to detect, differentiate and identify microorganisms are now widely used in many areas including food science [4]. A number of studies on development and introduction of such methods for meat product safety assessment is rapidly growing [5-9]. Some methods are based on polymerase chain reaction (PCR) such as specific-PCR, RAPD-PCR, PCR-DGGE, RFLP, AFLP, speciesspecific- PCR, real-time-PCR and multiplex-PCR [10-11]. From these, PCR with species-specific primers is probably the most widely used [12].
Two subspecies of Staphylococcus carnosus - Staphylococcus carnosus subsp. сarnosus and Staphylococcus carnosus subsp. utilis in starter cultures were successfully determined using real time PCR and High resolution Melting (HRM) analysis [13]. Naravaneni and Jamil [14] used PCR method to identify the food borne pathogens Salmonella and Escherichia coli.
The terminal restriction fragment length polymorphism (TRFLP) technique is considered as a rapid and reliable tool for microbial community fingerprinting. T-RFLP approach is based on restriction fragment analysis of a PCR amplified marker and automated sequencing gel technology. It allows to obtain the results with higher accuracy and resolution compared to other molecular technique [15]. In contrast to the real-time PCR which allows detection of only those microorganisms, for which the primers were selected, T-RFLP is used to detect all microorganisms including non-culturable. With an increasing demands on speed, ease of automation, accuracy and reproducibility of microbiological analysis, T-RFLP appears to be an attractive molecular approach to speed up the microbiological assays and provide access to essential information on microbiological quality and safety of food products.
Horse meat is a part of the traditional diet in Central Asia and in some European countries [16]. Although consumption of horse meat nowadays is not widespread [17], interest in horse meat is growing because of its high nutritional value [18-20] and lower environmentally harmful effects compared to beef [21]. Horse meat is consumed either cooked or processed (cured and fermented). Fermented horse meat sausage-like product Kazy is a habitual dish in several central Asian regions [22]. This is cured-raw product which does not undergo heat treatment during the manufacture and considered as safe by consumers. Indeed, no outbreaks due to Kazy were reported. Yet, the importance of fermented meats as a source of pathogens is well recognized [23]. To the best of our knowledge, only limited information is available on microbiological quality of horse meat. Gill and Landers (2005) demonstrated that the microbiological conditions of raw horse meat at different stages of processing are similar with these of beef. Alagić et al., [24] monitored changes in microflora during ripening of horsemeat sausages and showed prevalence of lactic acid bacteria, but also micrococci, yeast and fungi.
The aim of the present study was to characterize microbial community of the local fermented sausage-like product Kazy with respect to their microbiological safety. For this purpose, we used molecular genetic methods - T-RFLP and real-time PCR.
Local fermented sausage-like product Kazy was used in the study. The product contained horse meat, horse fat, salt, honey and garlic. To prepare the product, house meat was cut into 2–3 cm strips, followed by addition of fat and salt. After mixing, the product was cured for 24 hours. Then, honey and minced garlic was added. After mixing, sausage batter was filled into natural casings, settled at +4°C for 48 hours, gradually frozen to -10°C and ripened in well-ventilated environments for 1-1.5 month. For analysis of the product microflora on 5th day from the manufacturing date, 4 randomly selected products were cut and 3 samples (1g) were taken from the inner part of each product, homogenized in a ceramic mortar and pooled. The procedure was repeated 3 times. A total of 36 samples were collected, and 12 pooled samples were analysed.
The DNA was extracted with phenol/chloroform (1:1) solution and purified with the CTAB solution. Pooled sample (0.5 g) was transferred into an eppendorf tube (1.5 ml) with a screw cap. Then, 500 μl of buffer I (CTAB 2%; Tris-HCl 0.1M; EDTA-Na2 20 mM; NaCl 1.4 M; pH 8.5) and 0.5 g of glass beads (Helicon, Russia) were added to the sample. The sample was heated at 65°C for 15 min and homogenized on a personal Vortex V-1 (Biosan, Latvia) at 3000 rpm for 15 min; then, the heating process was repeated during 15 min. After that, the sample was centrifugedat 14000 rpm for 10 min (Mini Spin, Eppendorf, Germany) with 400 μl of phenol/chloroform mixture (1:1), the supernatant was then transferred to a new eppendorf tube and centrifuged again with 400 μl of chloroform. Afterwards, DNA was precipitated in a centrifuge at 14000 rpm with 400 μl of 96% ethanol in the presence of 0.3 Мsodium acetate (Helicon, Russia) and dissolved in 100 μl of TE buffer (Tris-HCl 10 mM; EDTA-Na2 1 mM) (Helicon, Russia).
The method has been adapted and applied to meat products. 16S rRNA genes were amplified using the primers 63F (CAGGCCTAACACATGCAAGTC) with a tag at the 5’-end (fluorophore D4-WellRed) and 1492R (TACGGHTACCTTGTTACGACTT). The mixture for PCR contained 10 pM of primers, 2.5 units of Taq| polymerase (Fermentas, USA), Х10 buffer for Taq polymerase (Fermentas, USA), 2 μl of 25 mM MgCl2 (Fermentas, USA),a mixture of deoxynucleotide triphosphates (dATP, dGTP, dCTP, dTTP at final concentration of 150 μM), 1 μl of DNA. The sample was adjusted to a volume of 20 μl with deionized water. PCR was performed in the amplifier MaxyGene (Axygen, USA) under the following conditions: 95 °С - 3 min, 35 cycles (95 °С - 30 s, 55 °С - 30 s, 72 °С - 60 s), 72 °С - 10 min.
An amplified fragment was isolated from the agarose gel using the 3М guanidine thiocyanate solution as following. An agarose block with the amplified fragments of DNA was removed from the agarose gel and placed into eppendorf tubes (1.5 ml). 100 μl of 3М guanidine isothiocyanate contained 20 mM EDTA-Na2, 10 mМ Tris-HCl (pH 6.8) and40 mg/ml of TritonX-100 (Helicon, Russia) was added to the block and heated to 65°С until the agarose block was completely dissolved. Then, the sample was mixed with 20μl of above described solution contained 40 mg/ml of DNA sorbent Silica (Helicon, Russia) and incubated at the room temperature for 10 min. Amplicon was precipitated with a sorbent in the centrifuge Mini Spin (Eppendorf, Germany) at 4000 rpm for 1 min. The sediment of silica with DNA was washed with 100 μl of solution contained 25% C2H5OH, 25% isopropanol, 100 mМ NaCl and 10 mМ TRIS-HCl, pH 8.0 (Helicon, Russia) and 70% ethanol. Then, the sediment was dried and the DNA was eluted in 100 μl of 10 mМTris-HCl buffer (pH 8.0) (Helicon, Russia) for 15 min at room temperature. Then, the solution was centrifuged at 14000 rpm for 3 min. and the purified DNA preparation was transferred into new tubes.
The PCR products were digested with 10 units of the restriction enzymes HaeIII, HhaI and MspI (Fermentas, USA) at 37°С for 2 hours. The restriction digests were then purified with ethanol in an amount of 38 μl in the presence of 1.5 μl of 3M sodium acetate solution and dissolved in 10 μl of SLS (Beckman Coulter, USA) with addition of 0.2 μl of marker with molecular weight of 600 bp (Beckman Coulter, USA). The fragments were analyzed by capillary electrophoresis (Frag4 program) with fluorescence detection and automated sequencer CEQ8000 (Beckman Coulter, USA).
Peak sizes and areas were determined on the Fragment Analysis software (Beckman Coulter, USA). Coefficient of variations (CV%) were below 5%. T-RFLP electrophoregrams were analyzed using Fragment Sorter (http://www.oardc.ohio-state.edu/trflpfragsort/index.php).
Determination of the total number of microorganisms and lactobacteria was performed by real-time PCR using the primers Eub338 5’-ACTCCTACGGGAGGCAGCAG-3’, Eub518 5’-ATTACCGCGGCTGCTGG-3’ (Syntol, Russia). The regime of PRC amplification was following: 95 °C - 3 min, (95 °C - 13 s, 63 °C - 13 s, 72 °C - 30 s) 40 cycles, 72 °C - 5 min. (Guo X. et al., 2008).
Quantification of bacteria of the genus Lactobacillus was carried out using the primers Lact-F (AGAGGTAGTAACTGGCCTTTA) и Lact-R (GCGGAAACCTCCCAACA) (Syntol, Russia). The regime of PRC amplification was as follows: 95 °C - 3 min, (95 °C - 30 s, 60 °C - 30 s, 72 °C - 1 min) 40 cycles, 72 °C - 5 min [27].
Amplification was carried out using «The reagent kit for performing real-time PCR with Taq DNA polymerase and antibodies inhibiting an activity of the enzyme in the presence of Eva Green dye» (LLC «NPO DNA-Technology») using the detecting amplifier DTlite (LLC «NPO DNA-Technology») according to manufacture instructions.
The statistical analysis of data obtained was carried out with the use of STATISTICA 6.0 Software Package, by application of the Student's t-test (differences at p<0.05 were considered statistically reliable). The mathematical treatment of the data including calculation of averages with standard errors (M ± m) was carried out.
Results and Discussion
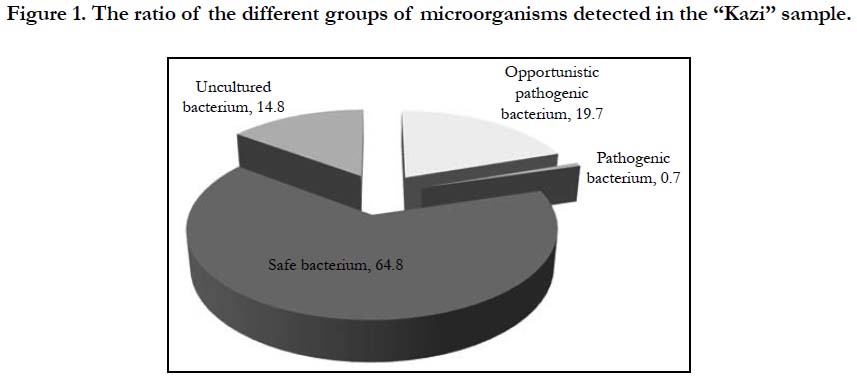
Microbiological analyses of fermented horse meat sausage-like product revealed an ordinary microbiological profile (Figure 1) with microflora mainly (approximately 80%) represented by safe and non-culturable microorganisms and to a lesser extent by conditionally pathogenic microorganisms.
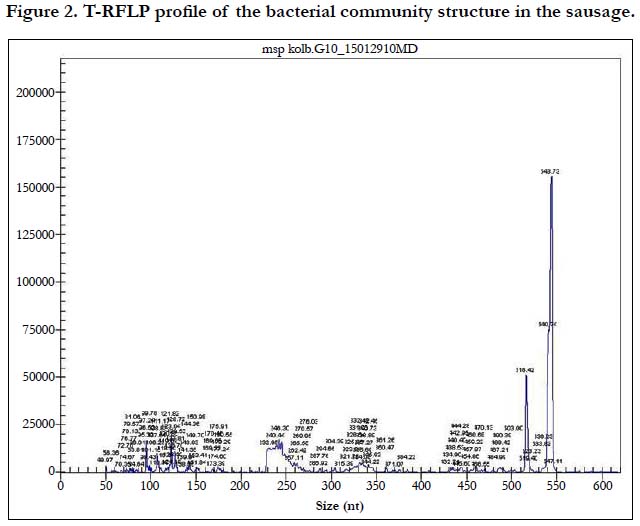
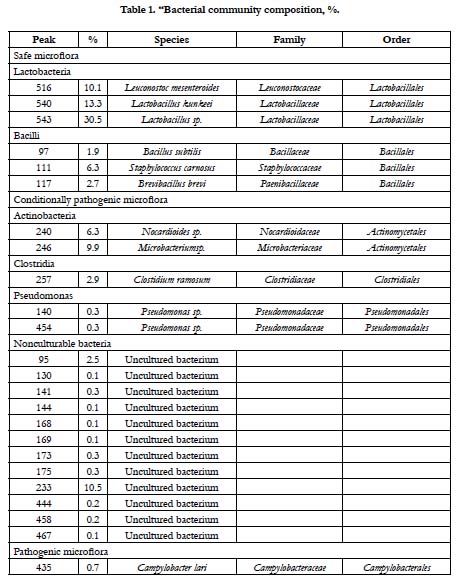
Pathogenic bacteria accounted for less than 1%. Conditionally pathogenic microflora was represented by actinobacteria Nocardioides spp. and Microbacterium spp., Clostidium ramosum and Pseudomonas, whereas coliform bacteria was not detected (Figure. 2, Table 1). Pathogenic microorganisms were represented by Сampylobacter lari. Nowadays, the acceptable limit for campylobacter counts in fermented sausages is not specified by the Russian Legislation. Campylobacter is considered to be the main cause of bacterial gastroenteritis in humans and is a significant public health burden.
Although epidemiological studies repeatedly suggest that the most significant Campylobacter pathogen species are C. jejuni and C. coli, C. lari was also recognized as a human pathogen [25, 26]. EFSA [27] estimated the losses due campylobacteriosis in the EU in the amount of € 2.4 billion a year. In this regard, there is growing demand for Campylobacter detection in foods which is challenging because most Campylobacter species are relatively metabolically inactive, which makes it difficult to identify them by traditional microbiological or biochemical methods. A major course of campylobacteriosis cases in humans is consumption of contaminated raw poultry meat [28], whereas contamination of horse meat with Campylobacter is uncommon [29, 30]. However, the presence of C. lari in the fermented horse meat sausage-like product in our study highlights the need for further research.
Non-culturable forms of bacteria accounted for 14.8% in the investigated samples. Non-culturable forms of bacteria are metabolically active, but lost ability to grow on routine media. Bacteria enters non-culturable form occurs under unfavorable environmental conditions, but becomes culturable when the unfavorable conditions are removed, and can pose health risks.
The results of the real-time PCR showed that lactic acid bacteria in fermented horse meat sausage-like product accounted for 4.00×104 ± 1.87×103 genomes/g or 50.6% of the total microbial number. The T-RFLP analysis of the same sample showed that lactic acid bacteria accounted for 53.9% of the total microbial number (7.90×104 ± 3.01×103 genomes/g). These similarities indicated that T-RFLP analysis can be successfully applied to characterize microflora in meat products and is an excellent tool for rapid and accurate identification of relevant bacteria.
In recent years, the application of this method has expanded into the area of meat safety. For example, T-RFLP-analysis was successfully used to study microbial spoilage of meat. Nieminen et al., [31] applied the T-RFLP method to examine psychrotrophic lactic acid bacteria (LAB) and Brochothrix thermosphacta communities in meat packed in modified atmosphere (MAP). Li et al., [32] characterized bacterial communities in beef spoiled after 10 days of aerobic storage at 4°C.
Rahkila et al., [33] isolated 222 psychrotrophic Lactococcus from the MAP-pork meat and identified with EcoRI and ClaI ribosomal patterns and phylogenetic analysis of 16S sequences, rpoA and pheS genes. Most microorganisms (N = 215) in that study were identified as Lactococcus piscium, while seven isolates identified as Lactococcus raffinolactis. The methods used have been shown to be reliable tools for Lactococcus species identification in meat.
T-RFLP-method is also used for seafood and fish bacterial community composition analysis. Tanaka et al., [34] described express system using 16S rDNA specified T-RFLP analysis to study microbial populations in fish. Database of terminal restriction fragments was constructed based on 102 bacterial strains of 53 species. T-RFLP system used gave results comparable to those obtained by the culture method in six fish samples with 71.4 to 92.3% compliance in 7 hours.
The results from the present and previous studies suggested that T-RFLP analysis is a rapid and suitable tool for monitoring microflora of fermented sausages or sausage-like products. Moreover, this method eliminates or minimize issues related to traditional culture-dependent methods.
Conclusions
Real-time PCR and T-RFPL approaches were successfully applied for analysis of microflora in fermented horse meat sausage-like product. We demonstrated that microflora in the fermented horse meat sausage-like product Kazy mainly consisted of safe and non-culturable bacteria (approximately 80%). The remaining part was represented by the conditionally pathogenic microflora, while the pathogenic microorganisms (Campylobacter) accounted only for .0.7% of total community. We suggest that real-time PCR and T-Real-time PCR and T-RFPL approaches were successfully applied for analysis of microflora in fermented horse meat sausage-like product. We demonstrated that microflora in the fermented horse meat sausage-like product Kazy mainly consisted of safe and non-culturable bacteria (approximately 80%). The remaining part was represented by the conditionally pathogenic microflora, while the pathogenic microorganisms (Campylobacter) accounted only for .0.7% of total community. We suggest that real-time PCR and TRFPL approaches can be effectively used for research purposes for detection of pathogenic and conditionally pathogenic microflora, and in inspection programs.
Financial Support
This work was supported by Russian Science Foundation (RSF) (project No. 16-16-10073).
References
- [1]. Casquete R, Martín A, Benito MJ, Ruiz‐Moyano S, Nevado FP, Córdoba MD. Impact of Pre‐Selected Autochthonous Starter Cultures on the Flavor Quality of Iberian Dry‐Fermented “Salchichón” Sausage with Different Ripening Processes. J Food Sci. 2011 Nov-Dec;76(9):S535-44. doi: 0.1111/j.1750-3841.2011.02425.x. PubMed PMID: 22416726.
- Danilović B, Joković N, Petrović L, Veljović K, Tolinački M, Savić D. The characterisation of lactic acid bacteria during the fermentation of an artisan Serbian sausage (Petrovská Klobása). Meat Sci. 2011 Aug;88(4):668-74. doi:10.1016/j.meatsci.2011.02.026. PubMed PMID: 21420794.
- Lahtinen SJ, Ahokoski H, Reinikainen JP, Gueimonde M, Nurmi J, Ouwehand AC, et al. Degradation of 16S rRNA and attributes of viability of viable but nonculturable probiotic bacteria. Lett Appl Microbiol. 2008 Jun;46(6):693-8. doi: 10.1111/j.1472-765X.2008.02374.x. PubMed PMID: 18444975.
- Ceuppens S, Li D, Uyttendaele M, Renault P, Ross P, Ranst MV, et al. Molecular methods in food safety microbiology: interpretation and implications of nucleic acid detection. Compr Rev Food Sci Food Saf. 2014 Jul;13(4):551-77.
- Botina SG, Tsygankov YD, Sukhodolets VV. Identification of industrial strains of lactic acid bacteria by methods of molecular genetic typing. Genetika. 2006 Dec;42(12):1621-35. PubMed PMID: 17326382.
- Kesmen Z, Yetiman AE, Gulluce A, Kacmaz N, Sagdic O, Cetin B, et al. Combination of culture-dependent and culture-independent molecular methods for the determination of lactic microbiota in sucuk. Int J Food Microbiol. 2012 Feb 15;153(3):428-35. doi: 10.1016/j.ijfoodmicro. 2011.12.008. PubMed PMID: 22209604.
- Fontana C, Gazzola SI, Cocconcelli PS, Vignolo G. Population structure and safety aspects of Enterococcus strains isolated from artisanal dry fermented sausages produced in Argentina. Lett Appl Microbiol. 2009 Sep;49(3):411-4. doi: 10.1111/j.1472-765X.2009.02675.x. PubMed PMID: 19627479.
- Wanangkarn A, Liu DC, Swetwiwathana A, Jindaprasert A, Phraephaisarn C, Chumnqoen W, et al. Lactic acid bacterial population dynamics during fermentation and storage of Thai fermented sausage according to restriction fragment length polymorphism analysis. Int J Food Microbiol. 2014 Sep 1;186:61-7. doi: 10.1016/j.ijfoodmicro.2014.06.015. PubMed PMID: 25005265.
- Rungrassamee W, Tosukhowong A, Klanchui A, Maibunkaew S, Plengvidhya V, Karoonuthaisiri N. Development of bacteria identification array to detect lactobacilli in Thai fermented sausage. J Microbiol Methods. 2012 Dec;91(3):341-53. doi: 10.1016/j.mimet.2012.09.016. PubMed PMID: 23022427.
- Gokulakrishnan P, Vergis J. Molecular Methods for Microbiological Quality Control of Meat and Meat Products: A Review. Crit Rev Food Sci Nutr. 2015;55(10):1315-9. doi: 10.1080/10408398.2012.691127. PubMed PMID: 24915322.
- Miller P, Liu X, McMullen LM. Microbiota of regular sodium and sodiumreduced ready-to-eat meat products obtained from the retail market. Can J Microbiol. 2015 Feb;61(2):150-4. doi: 10.1139/cjm-2014-0630. PubMed PMID: 25600580.
- Połka J, Rebecchi A, Pisacane V, Morelli L, Puglisi E. Bacterial diversity in typical Italian salami at different ripening stages as revealed by high-throughput sequencing of 16S rRNA amplicons. Food Microbiol. 2015 Apr;46:342-356. doi: 10.1016/j.fm.2014.08.023. PubMed PMID: 25475305.
- Chernukha IM, Minaev MY, Kurbakov KA, Bataeva DS. Detection and identification of S. carnosus in starter cultures using real time PCR and subsequent HRM analysis of amplification products. Procedia Food Sci. 2015 Jan 1;5:38-41.
- Naravaneni R, Jamil K. Rapid detection of food-borne pathogens by using molecular techniques. J Med Microbiol. 2005 Jan;54(Pt 1):51-4. PubMed PMID: 15591255.
- Liu WT, Marsh TL, Cheng H, Forney LJ. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997 Nov;63(11):4516-22. PubMed PMID: 9361437.
- Belaunzaran X, Bessa RJ, Lavín P, Mantecón AR, Kramer JK, Aldai N. Horse-meat for human consumption - Current research and future opportunities. Meat Sci. 2015 Oct;108:74-81. doi: 10.1016/j.meatsci.2015.05.006. PubMed PMID: 26047980.
- Cawthorn DM, Hoffman LC. Controversial cuisine: A global account of the demand, supply and acceptance of “unconventional” and “exotic” meats. Meat Sci. 2016 Oct;120:19-36. doi: 10.1016/j.meatsci.2016.04.017. PubMed PMID: 27155757.
- Badiani A, Nanni N, Gatta PP, Tolomelli B, Manfredini M. Nutrient Profile of Horsemeat1. J Food Compost Anal. 1997 Sep 1;10(3):254-69.
- Lee CE, Seong PN, Oh WY, Ko MS, Kim KI, Jeong JH. Nutritional characteristics of horsemeat in comparison with those of beef and pork. Nutr Res Pract. 2007 Spring;1(1):70-3. doi: 10.4162/nrp.2007.1.1.70. PubMed PMID: 20535389.
- Lorenzo JM, Sarriés MV, Tateo A, Polidori P, Franco D, Lanza M. Carcass characteristics, meat quality and nutritional value of horsemeat: A review. Meat Sci. 2014 Apr;96(4):1478-88. doi: 10.1016/j.meatsci.2013.12.006. PubMed PMID: 24423453.
- Moss AR, Jouany JP, Newbold J. Methane production by ruminants: its contribution to global warming. In: Annales de zootechnie. EDP Sci. 2000 May 1;49(3):231-253.
- Burçin Özvural E, Vural H. Fermented Sausages from Other Meats. Handbook of Fermented Meat and Poultry. 2014 Oct 2:339-43.
- Moore JE. Gastrointestinal outbreaks associated with fermented meats. Meat Sci. 2004 Aug;67(4):565-8. doi: 10.1016/j.meatsci.2003.12.009. PubMed PMID: 22061805.
- Alagić D, Kozačinski L, Filipović I, et al. The quality of fermented sausages of horse meat The quality of fermented sausages of horse meat during the three production seasons. Meso. 2008;10:200-203.
- Tauxe RV, Patton CM, Edmonds PA, Barrett TJ, Brenner DJ, Blake PA. Illness associated with Campylobacter laridis, a newly recognized Campylobacter species. J Clin Microbiol. 1985 Feb;21(2):222-5. PubMed PMID: 3972989.
- Werno AM, Klena JD, Shaw GM, Murdoch DR. Fatal case of Campylobacter lari prosthetic joint infection and bacteremia in an immunocompetent patient. J Clin Microbiol. 2002 Mar;40(3):1053-5. PubMed PMID: 11880437.
- Price KL, Totty HR, Lee HB, Utt MD, Fitzner GE, Yoon I, et al. Use of Saccharomyces cerevisiae fermentation product on growth performance and microbiota of weaned pigs during Salmonella infection. J Anim Sci. 2010 Dec;88(12):3896-908. doi: 10.2527/jas.2009-2728. PubMed PMID: 20656973.
- Konell K, Gelinsk MA, Benetti TM, Abrahão WM. Detection of thermophilic Campylobacter sp. in raw chicken sausages by methods ISO 10272: 2006 in Curitiba-Parana State-Brazil. Braz J Microbiol. 2015 Mar 4;45(4):1551-4. PubMed PMID: 25763066.
- Gill CO. Safety and storage stability of horse meat for human consumption. Meat Sci. 2005 Nov;71(3):506-13. doi: 10.1016/j.meatsci.2005.04.030. PubMed PMID: 22060926.
- Gill CO, Landers C. Microbiological condition of horse meat prepared at a North American packing plant, and control of the temperatures of product air freighted to Europe. Meat Sci. 2005 Mar;69(3):501-7. doi: 10.1016/j. meatsci.2004.09.005. PubMed PMID: 22062989.
- Nieminen TT, Vihavainen E, Paloranta A, Lehto J, Paulin L, Auvinen P, et al. Characterization of psychrotrophic bacterial communities in modified atmosphere-packed meat with terminal restriction fragment length polymorphism. Int J Food Microbiol. 2011 Jan 5;144(3):360-6. doi: 10.1016/j. ijfoodmicro.2010.10.018. PubMed PMID: 21093087.
- Li S, Zamaratskaia G, Roos S, Båth K, Meijer J, Borch E, et al. Inter-relationships between the metrics of instrumental meat color and microbial growth during aerobic storage of beef at 4° C. Acta Agric Scand A Anim Sci. 2015 Apr 3;65(2):97-106.
- Rahkila R, Nieminen T, Johansson P, Säde E, Björkroth J. Characterization and evaluation of the spoilage potential of Lactococcus piscium isolates from modified atmosphere packaged meat. Int J Food Microbiol. 2012 May 1;156(1):50-9. doi: 10.1016/j.ijfoodmicro.2012.02.022. PubMed PMID: 22445914.
- Tanaka Y, Takahashi H, Kitazawa N, Kimura B. Rapid estimation of microbial populations in fish samples by using terminal restriction fragment length polymorphism analysis of 16S rDNA. J Food Prot. 2010 Jan;73(1):104-13. PubMed PMID: 20051212.