Pharmacogenetic Associations with ADME Variants and Virologic Response to an Initial HAART Regimen in HIV-Infected Women
Pearce CL1,2*, Stram D2, Wiensch A1, Frasco MA2, Kono N2, Van Den Berg D2, Anastos K3, Cohen MH4, DeHovitz J5, Golub ET6, Tamraz B7, Liu C8, Mack WJ2
1 Department of Epidemiology, School of Public Health, University of Michigan, Ann Arbor, MI, USA.
2 Department of Preventive Medicine, Keck School of Medicine, University of Southern California, Los Angeles, CA, USA.
3 Department of Medicine, Montefiore Medical Center and Albert Einstein College of Medicine, Bronx, NY, USA.
4 Departments of Medicine, Stroger Hospital and Rush University, Chicago, IL, USA.
5 Department of Medicine and Community Health, SUNY Health Sciences Center, Brooklyn, NY, USA.
6 Department of Epidemiology, Johns Hopkins Bloomberg School of Public Health, Baltimore, MD, USA.
7 Department of Clinical Pharmacy, School of Pharmacy, University of California, San Francisco, CA, USA.
8 Department of Medicine, Georgetown University School of Medicine, District of Columbia, USA.
*Corresponding Author
Dr. Celeste Leigh Pearce
Department of Epidemiology,
University of Michigan School of Public Health, Ann Arbor, MI, USA
Tel: 734-764-3835
Fax: 734-764-3192
E-mail: lpearce@umich.edu
Received: July 19, 2017; Accepted: September 01, 2017; Published: September 05, 2017
Citation: Pearce CL, Stram D, Wiensch A, Frasco MA, Kono N, Van Den Berg D, et al., Pharmacogenetic Associations with ADME Variants and Virologic Response to an Initial HAART Regimen in HIV-Infected Women. Int J AIDS Res. 2017;4(3):154-160. doi: dx.doi.org/10.19070/2379-1586-1700031
Copyright: Pearce CL© 2017. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: Clinical response to highly active antiretroviral therapy (HAART) varies among different populations. A portion of this variability may be due to variation in genes involved in the absorption, distribution, metabolism, and excretion (ADME) of HAART.
Design: To identify genetic factors involved in virologic responses to HAART, 13 genes in ADME pathways were analyzed in a cohort of HIV-infected women on HAART. A total of 569 HIV-positive participants from the Women’s Interagency HIV Study who initiated HAART from 1994-2012 and had genotype data were included in these analyses.
Methods: Admixture maximum likelihood burden testing was used to evaluate gene-level associations between common genetic variation and virologic response (achieving <80 viral copies/mL) to HAART overall and with specific drug classes.
Results: Six statistically significant (P<0.05) gene-level burden tests were observed with response to specific regimen types. CYP2B6, CYP2C19 and CYP2C9 were significantly associated with response to protease inhibitor (PI)-based regimens. CYP2C9, ADH1A and UGT1A1 were significantly associated with response to triple nucleoside reverse transcriptase inhibitor (NRTI) treatment.
Conclusions: Although no genome-wide associations with virologic response to HAART overall were detected in this cohort of HIV-infected women, more statistically significant gene-level burden tests were observed than would be expected by chance (two and a half expected, six observed). It is likely that variation in one of the significant genes is associated with virologic response to certain HAART regimens. Further characterization of the genes associated with response to PI-based treatment is warranted.
2.Introduction
3.Methods
3.1. Women’s Interagency HIV Study (WIHS) Study Design
3.2. Inclusion Criteria for Current Analysis
3.3. Outcome
3.4. Genotyping
3.5. Covariates
3.6. Statistical Analysis
4.Results
5.Discussion
6.Acknwoledgements
7.References
Keywords
HIV; Women; HAART; Pharmacogenomics; ADME.
Introduction
Morbidity and mortality has declined markedly with the advent and widespread use of highly active antiretroviral therapy (HAART) for individuals with HIV infection [1, 2]. While the primary factor driving response to HAART is adherence, virologic response to HAART varies among adherent individuals and across populations [3]. Variation in genes that affect the absorption, distribution, metabolism, and excretion (ADME) of antiretrovirals (ARV) is thought to account for a portion of the variability in responses to treatment [4-6].
Several studies among HIV-infected individuals have evaluated virologic and immunologic responses, as well as adverse effects such as hypersensitivity reactions, neurotoxicity, hepatotoxicity, and hyperbilirubinemia, in relationship to genetic polymorphisms [4, 5]. The genes which have been most frequently examined in pharmacogenetic association studies in HIV disease include those in the cytochrome p450 family (CYP), as well as the ABCB1 and UGT1A1 genes. The protein products of these genes are reported to be involved in the metabolism and transport of two ARV drug classes, non-nucleoside reverse transcriptase inhibitors (NNRTIs) and protease inhibitors (PIs) [6-8].
Numerous studies have evaluated CYP-based genetic determinants of ARV exposure, but whether these polymorphisms translate to drug efficacy is controversial [9-19]. There has also been wide interest in the potential genetic influence of ABCB1 on HAART responses owing to the crucial role of P-glycoprotein (encoded by ABCB1) in the distribution and excretion of PIs and NNRTIs [7, 20-23]. However, variation in virologic responses to PI-containing regimens in HIV-infected patients who carry variant alleles of two independent ABCB1 polymorphisms has not been consistently observed [20-23]. Enhanced immunologic recovery in carriers of these ABCB1 variants has been observed in a recent analysis of HIV infected Chinese persons treated with PI-containing regimens (n=275) [24]. Loss of function alleles in the UGT1A1 gene are associated with drug-related hyperbilirubinemia; carriers of this variant treated with the PI atazanavir develop more severe hyperbilirubinemia, since atazanavir acts as an inhibitor to UGT1A1 [25].
Pregnane X receptor (NR1I2) is a nuclear receptor activated by several PIs, the nucleoside reverse transcriptase inhibitor (NRTI) abacavir, and the NNRTI efavirenz [26-28]. NR1I2 regulates the expression of CYP3A4, CYP2B6, and ABCB1, which mediate the metabolism and transport of several NNRTIs and PIs [26]. Pharmacokinetic associations between NR1I2 variation and the PI atazanavir have been reported [26-28]; however, pharmacogenetic associations with virologic response to HAART have not been explored. NRTIs are believed to be metabolized through alcohol dehydrogenase and the alcohol dehydrogenase 1A gene (ADH1A) has been examined for gene-environment interactions in the metabolism of abacavir and alcohol, although clinically significant interactions have not been identified [29].
Despite the potential for personalizing HAART regimens based on genetic predictors of response, pharmacogenetic studies of HAART responses to date have not provided conclusive evidence of associations. Previous pharmacogenetic studies in treated HIV-infected populations have focused on potential functional single nucelotide polymorphisms (SNPs) in genes that metabolize NNRTIs and PIs; the aim of this study was to comprehensively investigate genetic variation in ADME genes in the Women’s Interagency HIV Study (WIHS), a prospective multi-site observational study of multi-ethnic women infected with HIV, in relationship to virologic response. Associations between polymorphisms, derived from genomewide genotyping data, in CYP1A2, CYP2A6, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5, ADH1A, ABCB1, UGT1A1, and NR1I2 and virologic response to an intial HAART regimen were explored.
The WIHS is a prospective study of HIV-infected women and a comparison group of HIV-uninfected women who were recruited in four phases from six sites across the U.S.: Bronx/Manhattan, New York; Brooklyn, New York; Washington, D.C.; Los Angeles, CA; San Francisco/Bay Area, CA; and Chicago, IL [30, 31]. Data in these analyses were restricted to the first two recruitment waves and include HIV-infected women only. The original recruitment phase was conducted in 1994-1995 and the second phase of recruitment was done in 2001-2002. The HIV-infected women in the WIHS are representative of the racial/ethnic population of HIV-infected women in the U.S. [30, 31]. A more detailed description of the WIHS cohort has been published [30, 31].
Participants are seen for in-person visits every six months during which trained medical interviewers administer an extensive questionnaire, a clinical exam is performed, and biological samples are collected. Laboratory testing includes the measurement of HIV-1 viral loads (copies/ml) and CD4+ cell counts. At each biannual visit, detailed information on participants’ current treatment regimen, the ARV medications taken in the previous six months and medications used for comorbidities is obtained. Commencing at visit 9 (1998), data were collected on self-reported adherence to ARV medications as determined by a visual analog scale (100%, 95-99%, 75-95%, <75% over the past six months).
HIV-infected women who consented to genetic studies, initiated a regimen of three or more ARVs during study follow-up, maintained that regimen for a minimum of two consecutive study visits, had HIV RNA measurements at the visit immediately prior to, the visit of, and the visit immediately after initiating a three or more drug regimen were eligible for this pharmacogenetic study. Participants who were ARV therapy experienced prior to initiating a three drug regimen were included in the study only if at least two new ARV drugs were implemented as part of the initiated three drug HAART regimen. From October 1994 to September 2012, 1,307 WIHS participants initiated HAART. A total of 632 women were ineligible for this analysis: 138 had missing viral load data, 63 had undetectable viral loads at all relevant visits, 262 discontinued HAART during the study period, six had substantial missing covariate data, and for 163 participants only one new ARV was added to an existing regimen to achieve the definition of HAART. From a total of 675 eligible HAART initiators, 51 women were excluded because their treatment regimen did not fall within one of the three treatment categories (NRTI-, NNRTI-, and PI-based regimens) investigated here and 55 women were excluded due to missing genotype data, leaving a final analytic dataset of 569 WIHS participants.
HIV viral load levels were quantified with a nucleic acid sequence based amplification assay with a lower limit of quantification of 80 copies/ml. A positive virologic response (“responders”) was defined as achievement of an undetectable viral load at the visit during which the HAART regimen was first reported or at the visit subsequent to it, which corresponds to a maximum of 54 weeks of treatment. Participants not achieving an undetectable viral load at the visit of first reported HAART regimen use (corresponding to the previous six months) or the subsequent visit were classified as “non-responders”.
A subset of 13 genes was selected from whole-genome genotyping data generated from a cohort-wide GWAS utilizing the Omni Human 2.5M SNP Array (Illumina, San Diego CA). Genotypes for SNPs in genes involved in ADME pathways (e.g. CYP1A2, CYP2A6, CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP2E1, CYP3A4, CYP3A5, ADH1A, ABCB1, UGT1A1, and NR1I2) were extracted for participants eligible for this study. Genotype imputation was performed using IMPUTE2, with imputed genotypes generated by comparing our GWAS SNP data for each gene region with reference haplotype data from the 1000 Genomes Phase 1 variant set release (version 3). 90 Markov chain Monte Carlo iterations were used in the imputation process. Only genotypes with an imputation r2 of 0.50 or higher are included in the analysis. Coverage for each of the gene regions was calculated in each racial group by calculating pairwise r2 between our genotyped and imputed (imputation r2 ≥ 0.5 or greater, MAF ≥ 0.05) SNPs and every 1000 Genomes SNP in the region (MAF ≥ 0.05) using the 1000 Genomes samples of each respective racial group. SNPs that had an r2 of 0.95 or greater with our genotyped and imputed SNPs were considered "covered," and the proportion of SNPs covered in each region was calculated.
Genetic ancestry for the entire cohort was quantified by performing principal components analysis on ancestry informative markers included on the genome-wide panel. Genetic ancestry principal components (PC) covariates were used to control for population stratification in association models. Adherence data for this analysis were assessed at the visit at which the participant achieved an undetectable viral load (visit of HAART initiation or post-HAART initiation) since the adherence variable at this visit reflects HAART adherence in the six months preceding outcome ascertainment. Adherence data for non-responders was taken at the study endpoint, which was defined as the visit immediately after the HAART initiation visit. For modeling purposes, adherence was dichotomized as ≥ 95% or < 95% adherent. Type of HAART regimen used by the participants was categorized as dual NRTI/PI, dual NRTI/NNRTI, or ≥ 3 NRTIs.
The analysis includes women who self-identified as African-American, Hispanic, or Non-Hispanic White. Logistic regression was used to test associations between each SNP and achievement of an undetectable viral load. Genotypes were modeled as an ordinal variable, where common allele homozygotes, heterozygotes and minor allele homozygotes were coded as 0, 1, and 2, respectively.
Adherence, pre-HAART viral load and nadir CD4+ count prior to initiation were evaluated as potential confounders. Ultimately, since SNPs showed no association with self-reported adherence, pre-HAART viral load or nadir CD4+ cell counts, these variables were not included in the final model. Analyses were performed with adjustment for the top nine genetic ancestry PCs. Stratification on self-reported race/ethnicity did not affect the results.
There is evidence that ARV drug classes vary in their affinity for particular enzymes, and as such, the analyses were carried out with all regimens combined as well as separately by triple NRTI-alone regimens, NNRTI-containing or PI-containing regimens.
Given the lack of independence among our SNPs, an admixture maximum likelihood (AML) burden test was employed to evaluate whether a greater set of nominal associations per gene were observed than would be expected under the null distribution. Essentially, the AML method postulates that a given proportion of SNPs (α) within a set of SNPs is associated with the outcome and that the magnitude of each associated SNP’s effect will fall on a non-central χ2 distribution with non-centrality parameter η, a measure closely related to that SNP’s contribution to the genetic variance of the outcome variable. The α and η parameters are estimated using a pseudo-maximum likelihood method [32]. This burden test was run using the AMLcalc program (http://ccge.medschl.cam.ac.uk/software/aml) with 1000 simulations; and the maximum proportion of associated SNPs was set to 0.2 on the genotyped and imputed data. We also adjusted for the first nine ancestry principal components. A total of 52 burden tests were run; comprising four HAART analytic groups (all regimens and each of the three regimen types) with each of the 13 genes.
Results
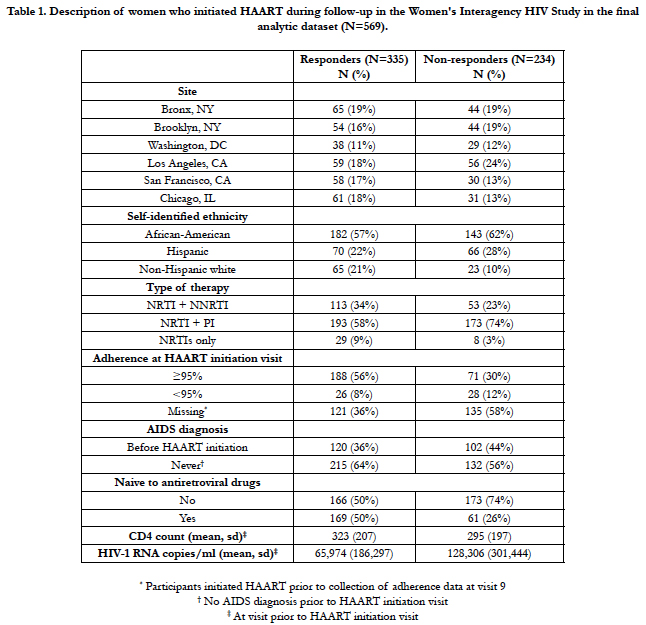
Table 1 summarizes the characteristics of HAART initiators from October, 1994 to September 2012 in the WIHS by virologic responder status. Of the 569 women included in the analysis, 59% were responders (n=335). Among the 313 (55%) women for whom self-reported adherence data were available, 12.1% of the responders reported <95% adherence compared to 28.3% of the non-responders.
Table 1. Description of women who initiated HAART during follow-up in the Women's Interagency HIV Study in the final analytic dataset (N=569).
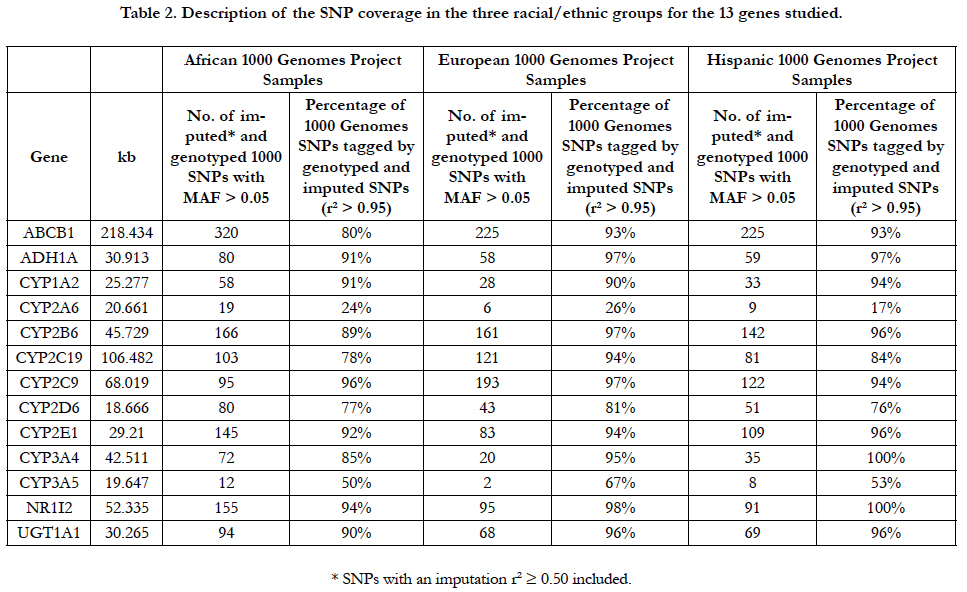
The 13 ADME genes evaluated in this analysis are shown in Table 2. The percentage of the genome covered in these regions by the genotyped and imputed SNPs by racial/ethnic group ranged from 17% to 100%. CYP2A6 and CYP3A5 were poorly covered in all of the racial/ethnic groups. Although the genotyping array used in this project was designed to provide good coverage for non-Whites, only six of the 13 genes were covered at 90% or higher for African-Americans compared to 10 and nine genes, respectively for Whites and Hispanics.
Table 2. Description of the SNP coverage in the three racial/ethnic groups for the 13 genes studied.
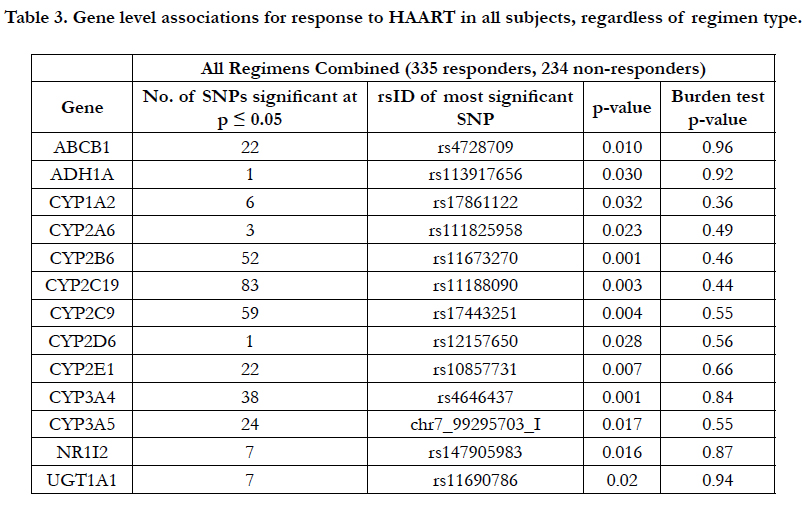
When considering the three types of HAART regimens (triple NRTIs, dual NRTI/PI, dual NRTI/NNRTI), individual associations at p<0.05 were observed in all 13 genes with virologic response (Table 3), but none of these reached genomewide significance. In order to further evaluate these nominally significant associations, gene-level burden testing analysis was conducted. None of the 13 burden tests was statistically significant (Table 3).
All genetic effects on virologic response were stratified based on type of regimen to test associations specific to treatment with triple NRTI-based, NNRTI-based and PI-based regimens.
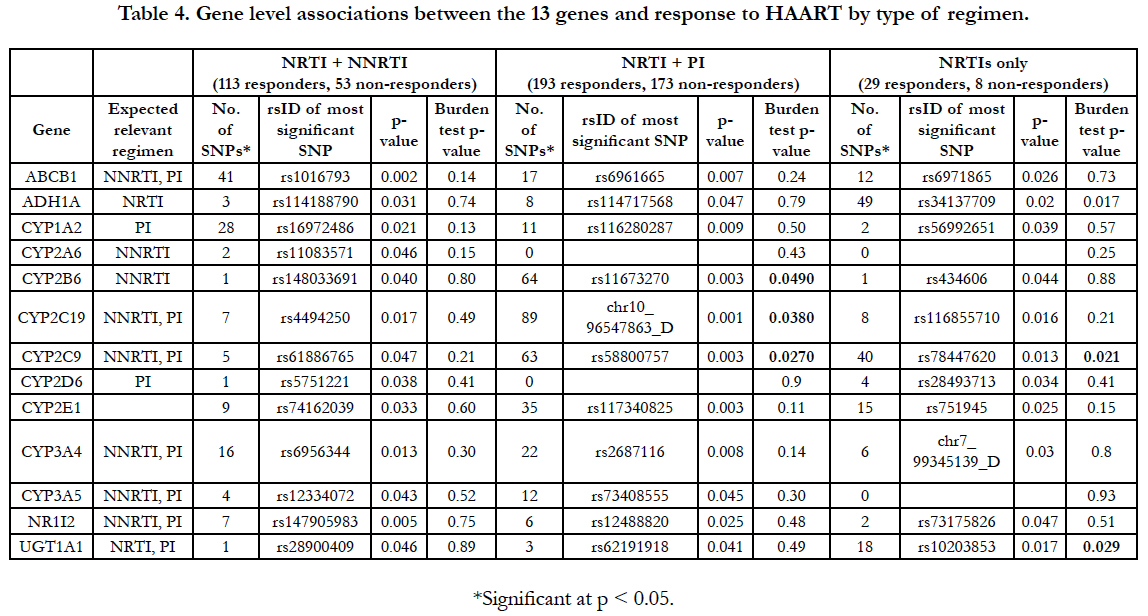
Associations between SNPs in these ADME genes and response to HAART were observed in almost all genes for the three regimen types. Again, no single association achieved genomewide significance in any of the three regimen types. Gene-level associations were assessed for each type of treatment regimen to evaluate evidence of association after taking into account all of the variation in the region and accounting for correlation between the SNPs. No significant (P<0.05) burden tests were observed for the NNRTI-based regimen group (Table 4). However, three significant burden tests were observed in each of the PI-based regimen and the triple NRTI-alone groups.
CYP2B6, CYP2C19, and CYP2C9 were significantly associated with virologic response to HAART at the gene level among women on a PI-based regimen (p-values 0.049, 0.038 and 0.027, respectively). CYP2C9 was also associated with response to HAART in the triple NRTI-alone group (p=0.021) as were ADH1A and UGT1A1 (p-values 0.017 and 0.029, respectively). By chance, 2.5 significant burden tests would be expected compared to six observed.
Discussion
We evaluated the relationship between variation in 13 genes involved in the absorption, distribution, metabolism and elimination of ARVs and virologic response to an initial HAART regimen and observed twice as many gene-level burden tests with p-values less than 0.05 compared to what was expected. This suggests that variation in CYP2B6, CYP2C19, and CYP2C9 are associated with virologic response to PI-based regimens and variation in CYP2C9, ADH1A and UGT1A1 are associated with responses to regimens containing three NRTIs.
The lack of association overall (Table 3) is not surprising given the expected class-specific effects of these genes. CYP2B6 is only known to be involved directly in the metabolism of NNRTIs, though its activity is inhibited by the pharmacoenhancer, ritonavir [33]. The burden p-value for CYP2B6 was 0.049 for the PI based regimen and this association may likely be due to chance. CYP2C19 and CYP2C9 (burden p values of 0.038 and 0.027, respectively) are both known to be involved in the metabolism of PIs [34, 35]. Genetic polymorphisms in both of these genes are known to markedly influence the clearance of other drugs [36].
ADH1A and UGT1A1 are the only genes known to be involved in the metabolism of NRTIs and the burden test p-values for these genes with triple NRTI regimens were 0.017 and 0.029, respectively. ADH1A and UGT1A1 are both involved in the metabolism of the NRTI abacavir [37]. SNPs in these regions could slow drug metabolism, possibly resulting in higher serum concentrations and more effective responses. CYP2C9 was also associated with NRTI-only treatment (burden test p value 0.021); as this gene was not previously known to be involved with NRTI metabolism, this could be a chance association. NRTIonly regimens are no longer recommended, but these results do provide insight into the metabolism of these drugs.
The major limitation of this study is the small sample size resulting in limited power to detect modest effects. Burden testing is a useful tool in this context to determine whether there is value in further follow-up of observed associations. Although some of the burden test results in our study may still be due to chance, the observation of six significant results suggests that one of these regions might be associated with virologic response to HAART and further study is warranted. The next steps in evaluating these associations are complicated by the fact that a region and not a SNP has been implicated. Thus for replication and extension of our findings, dense coverage of the regions would be needed and, optimally the population would also be female, as gender-specific effects cannot be ruled out. Fine mapping of the regions in the WIHS population that included rare variants is another option; if the true causal allele(s) are associated with a stronger effect, it might be possible to identify which of these six regions is truly associated with virologic response to HAART. In addition, some of these regions have poor coverage, particularly in the African American populations (Table 2), which likely limits the ability to detect significant associations for this group.
Though no genome wide assocations were detected, several gene level statistically significant associations were identified. Despite the low power of this study, at least one of these associations is likely to be true and not due to chance alone. Defining genetic factors that impact virologic response could lead to improvements in individualizing treatment for HIV-positive patients through genetic screening. Further studies are needed to more finely map these genetic regions harboring putative variants associated with virologic responses to specific HAART regimens.
Acknowledgements
Data in this manuscript were collected by the Women’s Interagency HIV Study (WIHS). The contents of this publication are solely the responsibility of the authors and do not represent the official views of the National Institutes of Health (NIH). WIHS (Principal Investigators): UAB-MS WIHS (Mirjam- Colette Kempf, and Deborah Konkle-Parker), U01-AI-103401; Atlanta WIHS (Ighovwerha Ofotokun and Gina Wingood), U01-AI-103408; Bronx WIHS (Kathryn Anastos), U01-AI-035004; Brooklyn WIHS (Howard Minkoff and Deborah Gustafson), U01- AI-031834; Chicago WIHS (Mardge Cohen and Audrey French), U01-AI-034993; Metropolitan Washington WIHS (Seble Kassaye), U01-AI-034994; Miami WIHS (Margaret Fischl and Lisa Metsch), U01-AI-103397; UNC WIHS (Adaora Adimora), U01-AI-103390; Connie Wofsy Women’s HIV Study, Northern California (Ruth Greenblatt, Bradley Aouizerat, and Phyllis Tien), U01-AI-034989; WIHS Data Management and Analysis Center (Stephen Gange and Elizabeth Golub), U01-AI-042590; Southern California WIHS (Joel Milam), U01-HD-032632 (WIHS I – WIHS IV). The WIHS is funded primarily by the National Institute of Allergy and Infectious Diseases (NIAID), with additional cofunding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), the National Cancer Institute (NCI), the National Institute on Drug Abuse (NIDA), and the National Institute on Mental Health (NIMH). Targeted supplemental funding for specific projects is also provided by the National Institute of Dental and Craniofacial Research (NIDCR), the National Institute on Alcohol Abuse and Alcoholism (NIAAA), the National Institute on Deafness and other Communication Disorders (NIDCD), and the NIH Office of Research on Women’s Health. WIHS data collection is also supported by UL1-TR000004 (UCSF CTSA), UL1-TR000454 (Atlanta CTSA), and P30-AI-050410.
References
- Palella FJ, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338(13):853-860.
- Gange SJ, Barrón Y, Greenblatt RM, Anastos K, Minkoff H,Young M, et al. Effectiveness of highly active antiretroviral therapy among HIV-1 infected women. J Epidemiol Community Health. 2002 Feb;56(2):153-159.
- Esposito A, Floridia M, d'Ettorre G, Pastori D, Fantauzzi A, Massetti P, et al. Rate and determinants of treatment response to different antiretroviral combination strategies in subjects presenting at HIV-1 diagnosis with advanced disease. BMC Infect Dis. 2011 Dec 14;11:341.
- Owen A, Khoo SH. Pharmacogenetics of antiretroviral agents. Curr Opin HIV AIDS. 2008 May;3(3):288-295.
- Tozzi V. Pharmacogenetics of antiretrovirals. Antiviral Res. 2010 Jan;85(1):190-200.
- Michaud V, Bar-Magen T, Turgeon J, Flockhart D, Desta Z, Wainberg MA. The dual role of pharmacogenetics in HIV treatment: mutations and polymorphisms regulating antiretroviral drug resistance and disposition. Pharmacol Rev. 2012 Jul;64(3):803-833.
- Weiss J, Weis N, Ketabi-Kiyanvash N, Storch CH, Haefeli WE. Comparison of the induction of P-glycoprotein activity by nucleotide, nucleoside, and non-nucleoside reverse transcriptase inhibitors. Eur J Pharmacol. 2008 Jan;579(1-3):104-109.
- Storch CH, Theile D, Lindenmaier H, Haefeli WE, Weiss J. Comparison of the inhibitory activity of anti-HIV drugs on P-glycoprotein. Biochem Pharmacol. 2007 May 15;73(10):1573-1581.
- Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genomics. 2005 Jan;15(1):1-5.
- Haas DW, Smeaton LM, Shafer RW, Robbins GK, Morse GD, Labbe L, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing Efavirenz and/or Nelfinavir: an Adult Aids Clinical Trials Group Study. J Infect Dis. 2005 Dec 1;192(11):1931-1942.
- Fellay J, Marzolini C, Decosterd L, Golay KP, Baumann P, Buclin T, et al. Variations of CYP3A activity induced by antiretroviral treatment in HIV-1 infected patients. Eur J Clin Pharmacol. 2005;60(12):865-873.
- Anderson PL, Lamba J, Aquilante CL, Schuetz E, Fletcher CV. Pharmacogenetic characteristics of indinavir, zidovudine, and lamivudine therapy in HIV-infected adults: a pilot study. J Acquir Immune Defic Syndr. 2006 Aug;42(4):441-449.
- Motsinger AA, Ritchie MD, Shafer RW, Robbins GK, Morse GD, et al. Multilocus genetic interactions and response to efavirenz-containing regimens: an adult AIDS clinical trials group study. Pharmacogenet Genomics. 2006;16(11):837-845.
- Rotger M, Tegude H, Colombo S, Cavassini M, Furrer H, Decosterd L, et al. Predictive value of known and novel alleles of CYP2B6 for efavirenz plasma concentrations in HIV-infected individuals. Clin Pharmacol Ther. 2007 Apr;81(4):557-566.
- Saitoh A, Sarles E, Capparelli E, Aweeka F, Kovacs A, Fenton T, et al. CYP2B6 genetic variants are associated with nevirapine pharmacokinetics and clinical response in HIV-1-infected children. AIDS. 2007 Oct 18;21(16):2191-2199.
- Haas DW, Gebretsadik T, Mayo G, Menon UN, Acosta EP, Shintani A, et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African americans. J Infect Dis. 2009 Mar 15;199(6):872-880.
- Saitoh A, Capparelli E, Aweeka F, Sarles E, Singh KK, Kovacs A et al. CYP2C19 genetic variants affect nelfinavir pharmacokinetics and virologic response in HIV-1-infected children receiving highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2010 Jul 1;54(3):285-289.
- Carr DF, la Porte CJ, Pirmohamed M, Owen A, Cortes CP. Haplotype structure of CYP2B6 and association with plasma efavirenz concentrations in a Chilean HIV cohort. J Antimicrob Chemother. 2010 sep;65(9):1889-1893.
- Gandhi M, Greenblatt RM, Bacchetti P, Jin C, Huang Y, Anastos K, et al. A single-nucleotide polymorphism in CYP2B6 leads to >3-fold increases in efavirenz concentrations in plasma and hair among HIV-infected women. J Infect Dis. 2012 Nov 1;206(9):1453-1461.
- Fellay J, Marzolini C, Meaden ER, Back DJ, Buclin T, Chave JP, et al. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet. 2002 Jan 5; 359(9300):30-36.
- Brumme ZL, Dong WW, Chan KJ, Hogg RS, Montaner JS, Harrigan PR, et al. Influence of polymorphisms within the CX3CR1 and MDR-1 genes on initial antiretroviral therapy response. AIDS. 2003 Jan 24;17(2):201-208.
- Haas DW, Wu H, Li H, Bosch RJ, Lederman MM, Benson C, et al. MDR1 gene polymorphisms and phase 1 viral decay during HIV-1 infection: an adult AIDS Clinical Trials Group study. J Acquir Immune Defic Syndr. 2003 Nov; 34(3):295-298.
- Saitoh A, Singh KK, Powell CA, Fenton T, Fletcher CV, Starr S, et al. An MDR1-3435 variant is associated with higher plasma nelfinavir levels and more rapid virologic response in HIV-1 infected children. AIDS. 2005 Mar 4;19(4):371-380.
- Zhu P, Zhu Q, Zhang Y, Ma X, Li Z, et al. (2013) ABCB1 variation and treatment response in AIDS patients: initial results of the Henan cohort. PLoS One. 2013; 8(1):e55197.
- Rotger M, Taffe P, Bleiber G, Gunthard HF, Furrer H, Vernazza P, et al. Gilbert syndrome and the development of antiretroviral therapy-associated hyperbilirubinemia. J Infect Dis. 2005 Oct;192(8):1381-1386.
- Svärd J, Spiers JP, Mulcahy F, Hennessy M. Nuclear receptor-mediated induction of CYP450 by antiretrovirals: functional consequences of NR1I2 (PXR) polymorphisms and differential prevalence in whites and sub-Saharan Africans. J Acquir Immune Defic Syndr. 2010 dec 15;55(5):536-549.
- Siccardi M, D'Avolio A, Baietto L, Gibbons S, Sciandra M, Bonora S, et al. Association of a single-nucleotide polymorphism in the pregnane X receptor (PXR 63396C-->T) with reduced concentrations of unboosted atazanavir. Clin Infect Dis. 2008 Nov1;47(9):1222-1225.
- Schipani A, Siccardi M, D'Avolio A, Baietto L, Simiele M, Bonora S, et al. Population pharmacokinetic modeling of the association between 63396C- >T pregnane X receptor polymorphism and unboosted atazanavir clearance. Antimicrob Agents Chemother. 2010 Dec;54(12):5242-5250.
- McDowell JA, Chittick GE, Stevens CP, Edwards KD, Stein DS. Pharmacokinetic interaction of abacavir (1592U89) and ethanol in human immunodeficiency virus-infected adults. Antimicrob Agents Chemother. 2000 Jun;44(6):1686-1690.
- Barkan SE, Melnick SL, Preston-Martin S, Weber K, Kalish LA, et al. The Women's Interagency HIV Study. WIHS Collaborative Study Group. Epidemiology. 1998 Mar; 9(2):117-125.
- Bacon MC, von Wyl V, Alden C, Sharp G, Robison E, et al. The Women's Interagency HIV Study: an observational cohort brings clinical sciences to the bench. Clin Diagn Lab Immunol. 2005 Sep;12(9):1013-1019.
- Tyrer J, Pharoah PD, Easton DF. The admixture maximum likelihood test: a novel experiment-wise test of association between disease and multiple SNPs. Genet Epidemiol. 2006 Nov;30(7):636-643.
- Lin HL, D'Agostino J, Kenaan C, Calinski D, Hollenberg PF. The effect of ritonavir on human CYP2B6 catalytic activity: heme modification contributes to the mechanism-based inactivation of CYP2B6 and CYP3A4 by ritonavir. Drug Metab Dispos. 2013 Oct;41(10):1813-1824.
- Damle BD, Uderman H, Biswas P, Crownover P, Lin C, et al. Influence of CYP2C19 polymorphism on the pharmacokinetics of nelfinavir and its active metabolite. Br J Clin Pharmacol. 2009 Nov;68(5):682-689.
- Barry M, Gibbons S, Back D, Mulcahy F. Protease inhibitors in patients with HIV disease. Clinically important pharmacokinetic considerations. Clin Pharmacokinet. 1997 Mar;32(3):194-209.
- Scordo MG, Pengo V, Spina E, Dahl ML, Gusella M, Padrini R. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on warfarin maintenance dose and metabolic clearance. Clin Pharmacol Ther. 2002 Dec;72(6):702-710.
- Barbarino JM, Kroetz DL, Altman RB, Klein TE. PharmGKB summary: abacavir pathway. Pharmacogenet Genomics. 2014 May;24(5):276-282.