Use of Tropical Strains: Aspergillus vadensis and Aspergillus oryzae as Producers of α-Amylases in Biotechnological Practice
Adejuwon AO1,2*, Tsygankova VA3
1 Department of Microbiology, Faculty of Sciences, Lead City University, Ibadan, Nigeria.
2 Department of Microbiology, Obafemi Awolowo University, Ile-Ife, Nigeria.
3 Department of Chemistry of Bioactive Nitrogen-Containing Heterocyclic Compounds, Institute of Bioorganic Chemistry and Petrochemistry of the National Academy of Sciences of Ukraine, Murmanskaya Str., Kyiv, Ukraine.
*Corresponding Author
Adekunle Odunayo Adejuwon
Department of Microbiology, Faculty of Sciences,
Lead City University, Ibadan, Nigeria.
E-mail: ao_adejuwon@yahoo.ca
Received: May 02, 2017; Accepted: June 12, 2017; Published: June 13, 2017
Citation: Adejuwon AO, Tsygankova VA (2017) Use of Tropical Strains: Aspergillus vadensis and Aspergillus oryzae as Producers of α-Amylases in Biotechnological Practice. Int J Med Biotechnol Genetics. 5(2), 57-63. doi: dx.doi.org/10.19070/2379-1020-1700010
Copyright: Adejuwon AO© 2017. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
Enzymes especially α-amylases are imported into Nigeria, West Africa in tons from the United States of America, Asia and East Europe, cumulating into annual national debts because of the dwindling exchange rate in foreign funds. There is avenue for the production of this enzyme locally in the tropics and even in the temperate zones of the world since production of this enzyme is advocated. In our study the isolates were obtained from deteriorated barley (Hordeum vulgare L.) plants grown locally in Jos, Plateau State, Nigeria. Tropical strains of Aspergillus vadensis and Aspergillus oryzae were isolated from deteriorated barley seeds placed onto potato dextrose agar slants and plates at 25°C. Sterile barley-seed medium (1% w/v) and a defined growth medium consisting MgSO4.7H20, K2HPO4, KH2PO4, aspartic acid, biotin, thiamine and FeSO4.7H2O with a nitrogen source and a carbon source for fungal growth were inoculated with spore suspensions of approximately 5 x 106 spores per ml of one hundred and twenty-hour-old cultures of each isolate. Incubation was at 25°C. The carbon source was varied and was independently starch, maltose, sucrose, lactose, glucose or galactose when the nitrogen source was potassium nitrate. The nitrogen source was also varied and was independently ammonium chloride, urea, potassium nitrate, ammonium sulphate, glycine, sodium nitrate, tryptone or peptone, when the carbon source in the growth medium was starch. Extracellular proteins produced within a ten day incubation period were extracted and assayed for α-amylase activity using the Pfueller and Elliott (1969) method. Extracellular proteins with expression of α-amylase activities were produced by the tropical fungal strains: Aspergillus vadensis and Aspergillus oryzae grown on the barley-seed medium and the defined growth medium used in this investigation within a period of ten days at 25°C. For the production of the most active α-amylases, the best carbon sources were maltose and glucose while the best nitrogen sources were ammonium chloride and ammonium sulphate when fungal strain Aspergillus vadensis was used as an inoculum. However, the best carbon sources were barley-seed medium, starch and sucrose while the best nitrogen sources were urea, glycine and sodium nitrate when the fungal strain Aspergillus oryzae was used as an inoculum. The exploration of this innovative is an a venue for the industrial production of α-amylases. This is encouraged in tropical Nigeria, West Africa, Ukraine, and East Europe.
2.Introduction
3.Materials and Methods
3.1 Source and Identification of Isolates
3.2 Culture Conditions and Inocula
3.3 Barley Seeds as Source of Carbon
3.4 Extraction of Enzyme
3.5 Enzyme Assay
4.Results
5.Discussion
6.Acknowledgements
7.References
Keywords
α-Amylase; Aspergillus vadensis; Aspergillus oryzae; Barley (Hordeum Vulgare L.); Growth Medium..
Introduction
α-Amylases are starch degrading enzymes that catalyze the hydrolysis of α-1,4-O-glycosidic bonds in polysaccharides with the retention of α-anomeric configuration in the products. Most of the α-amylases are metalloenzymes which require calcium ions (Ca2+) for their activity, structural integrity and stability [1]. Amylases are one of the most important industrial enzymes that have a wide variety of applications ranging from conversion of starch to sugar syrups, to the production of cyclodextrins for the pharmaceutical industry. These enzymes account for about 30% of the world’s enzyme production [2]. The α-amylase family can roughly be divided into two groups: the starch hydrolyzing enzymes and the starch modifying or transglycosylating enzymes [3]. It has been variously documented that the fungi are capable of production of enzymes including the α-amylases [4, 5].
An expression vector containing the Taka-amylase A gene from Aspergillus oryzae, which was fused to the strong promoter and signal peptide sequence of the cellobiohydrolase 1 gene (cbh 1) of Trichoderma viride and the hygromycin B resistance gene was used to transform protoplasts of Trichoderma viride [6]. Cloning of genes of amylase from rumen microorganisms mainly Escherichia coli using molecular biological techniques has been made possible in new attempts in the field of rumen microbiology [7]. Amylase produced by Saccharomyces cerevisaee when cocultured with Monascus isolate was effector of significant morphological changes including enhanced cell growth and pigment production in Monascus [8]. Studies carried out by Skovgaard and Rosendahl [9] revealed the production of amylases by Fusarium oxysporum from different habitats in culture media. According to Legin et al., [10], Thermococcus hydrothermalis and Thermococcus fumicolans isolated from deep sea were amylase producers. Sporothichum thermophile Apinis produce amylase with activity reaching approximately 45.2 M-Units after 5 days of growth [11]. Hayashi and Nozaki [12] reported the isolation and characterization of gibbestatin B, an inhibitor of gibberellin-induced expression of alpha-amylase and gibbestatin C from Streptomycetaes. Amylase producers, Vibrio and Aeromonas found in the mid gut of thalassinidea mud shrimp (Upogebia stellata) are likely to play significant roles in the digestion of gut content [13]. An amylolytic activity that converts soluble starch to α, α-trehalose (trehalose) was found in the cell homogenate of hyperthermophilic, acidophilic archaeum Sulfobolus sulfataricus KM1 [14]. α-Amylase with activity reaching 289 U/L was detected in a cell extract of Thermcoccus hydrothermalis grown in culture medium after 24h [15]. Ogawa et al., [16] reported the inactivation of alpha amylase by basic dyes. Douglas et al., [17] in their study demonstrated the detection of a noncovalent proteinprotein complex between alpha amylase and its microbial inhibitor tendamistat using ESI-MS. Two amylolytic enzymes, purified using affinity and ion-exchange chromatography were isolated from a thermophilic strain of Bacillus licheniformis [18]. A mutant from the co-culture of Aspergillus foetidus and Rhizopus oryzae was sensitive to tetracycline and was an over producer of amylase when compared with the wild strain [19]. Transformed strains of Saccharomyces kluyveri (investigated under aerobic glucose-limited conditions) and Saccharomyces cerevisaee secreted alpha-amylase with similar yields during continuous cultivations. Saccharomyces kluyveri was able to produce α-amylase concentrations as high as 320mg/l in a fed-batch cultivation [20]. A variant of Saccharomyces cerevisiae pep4 strain 20B12 with improved oligotrophic proliferation and cell survival is able to secrete heterologous mouse alpha-amylase [21]. The RegA protein, a Clostridium acetobutylicum regulator gene product affecting amylase production in Bacillus subtilis inhibited the degradation of starch by Clostridium acetobutylicum [22]. The application of dual promoters in recombinant DNA technology increased the productivity of α-amylase from Bacillus amyloliquefaciens [23]. Studies on the nucleotide sequence of the coding and regulatory regions of the alpha amylase gene (ama1) of Streptomyces limosus have revealed that the amino-terminal sequence of the extracellular enzyme contains a typical signal peptide preceding the mature form of the α-amylase [24]. Sequencing upstream of Streptococcus mutans gene with homology to a CcpA gene, regM, revealed an open reading frame, named amy, with homology to genes encoding α-amylase [25]. Transacting regulatory protein(s) interact with region III in the promoter regions of the amylase-encoding genes amy B, gla A and agd A of Aspergillus oryzae that are common to these amylase genes [26]. In a study involving the natural viability of the thermophilic culture of Bacillus diasticus, the population of the cells of the isolate was observed to be genetically heterologous with the bulk of the cells (75%) showing high activity of amylase biosynthesis [27].
This work is aimed to optimization of composition of growth medium to increase production of extracellular α-amylases by two fungal strains: Aspergillus vadensis and Aspegillus oryzae isolated from deteriorated seeds of barley (Hordeum vulgare L.).
The tropical fungal strains Aspergillus vadensis and Aspegillus oryzae for this research were isolated from deteriorated seeds of barley (Hordeum vulgare L.) grown in Jos, Plateau State, Nigeria. They were identified at the Seed Health Unit of the International Institute of Tropical Agriculture (IITA), Ibadan, Nigeria using standard fungi identification techniques [6]. Identity of fungi was further confirmed by genetic DNA sequencing using Polymerase Chain Reaction (PCR) at the Bioscience Laboratory of the International Institute of Tropical Agriculture (IITA), Ibadan, Nigeria. The isolates were cultured on potato dextrose agar slants and plates.
The tropical fungal strains Aspergillus vadensis and Aspergillus oryzae isolated from deteriorated barley seeds were grown in a defined growth medium with varying nitrogen and carbon sources of fungal growth. The isolates were also grown on barley-seed medium used as carbon source of fungal growth. The isolates Aspergillus vadensis and Aspergillus oryzae were cultured and maintained on potato dextrose agar slants and plates. Each fungus was sub-cultured on the test tubes of the same medium and incubated at 25°C. The one hundred and twenty-hour-old cultures of each isolate were used in this investigation. According to the modified method of Olutiola and Ayres [29], cultures were grown in a defined basal medium of the following composition: MgSO4.7H20 (0.1 g), K2HPO4 (2 g), KH2PO4 (0.5 g), aspartic acid (0.1 g), biotin (0.005 mg), thiamine (0.005 mg) and FeSO4.7H2O (1 mg) with a nitrogen source (9.9 g) and a carbon (10 g) source (Sigma) per 1 litre of distilled water. When the carbon source used was varied, the nitrogen source was potassium nitrate (9.9 g per litre of medium). The carbon sources were independently starch, maltose, sucrose, lactose, glucose and galactose (10g per litre of medium). When the nitrogen source used was varied, the carbon source was starch (10g per litre of medium). The nitrogen sources were independently ammonium chloride, urea, potassium nitrate, ammonium sulphate, glycine, sodium nitrate, tryptone and peptone (9.9 g per litre of medium). Conical flasks (250 ml) containing 100 ml growth medium were inoculated with 1 ml of an aqueous spore suspension containing approximately 5 x 106 spores per 1 ml of isolate. Spores were counted using the Neubauer counting chamber [30, 31]. Experimental and control flasks were incubated without shaking at 25°C [32]. Protein content of the inoculated medium was determined using the Lowry et al., [33] method.
The healthy Barley (Hordeum vulgare L.) seeds were obtained from the Jos International Brewery, Jos, Plateau State, Nigeria. They were weighed, soaked in distilled water (1% w/v) and then sterilized by autoclaving at 15 psi (121°C) for 15 minutes. This temperature ensures inactivation of both endogenous α-amylase and α-amylase inhibitor in the barley seeds but with care not to destroy the carbohydrate content of the seeds [34]. The each 100 ml of the barley-seed medium contained in conical flasks (volume of 250 ml) was inoculated with 1 ml of aqueous spore suspension containing approximately 5 x 106 spores per 1 ml of one hundred and twenty-hour-old cultures of isolate. Incubation was at 25°C.
On a daily basis, the contents of each flask were carefully filtered through glass fibre filter paper (Whatman GF/A). The protein content of the filtrates was determined [33]. The filtrates were also assayed for α-amylase activity [35].
α-Amylase activity was determined using the method of Pfueller and Elliott [35]. Reaction mixtures consisted 2 ml of 0.2% (w/v) starch in 0.2 M citrate phosphate buffer, pH 6.0 as substrate and 0.5 ml of enzyme. Controls consisted only 2 ml of the prepared substrate. The contents of both experimental and control tubes were incubated at 35°C for 20 min. The reaction in each tube was terminated with 3 ml of 1 N HCl. 0.5 ml enzyme was then added to the control tubes. The 2ml mixture of each experimental and control was transferred to new sets of test tubes. The 3ml of 0.1 N HCl was added into the contents of each test tube after which 0.1 ml of iodine solution was added. Optical density readings were taken at 670nm. One unit of α-amylase activity was defined as the amount of α-amylase which produced 0.1 percent reduction in the intensity of the blue colour of starchiodine complex under conditions of the assay.
Results
The isolates of two tropical fungal strains: Aspergillus vadensis and Aspergillus oryzae obtained from deteriorated Barley (Hordeum vulgare L.) seeds grew and exhibited α-amylase activities in both barley-seed medium and the defined growth medium used in this investigation at the temperature of 25°C (Figure 1).
Figure 1. The Isolates of Two Tropical fungal strains: A - Aspergillus oryzae and B - Aspergillus vadensis Inoculated on Potato Dextrose Agar Plates.
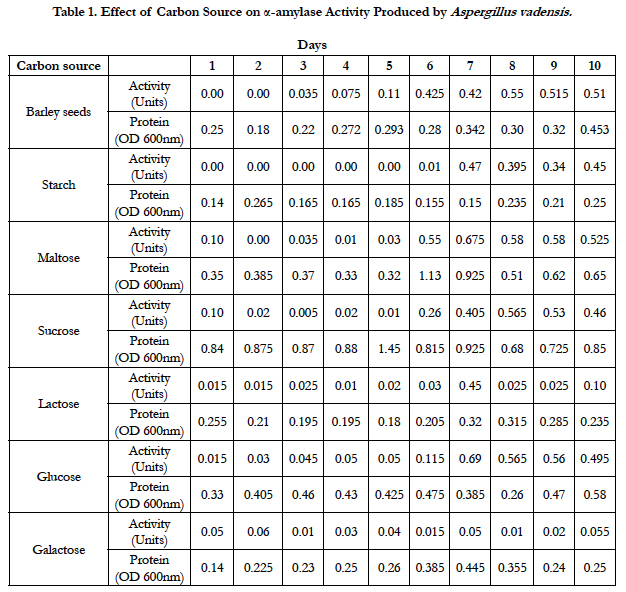
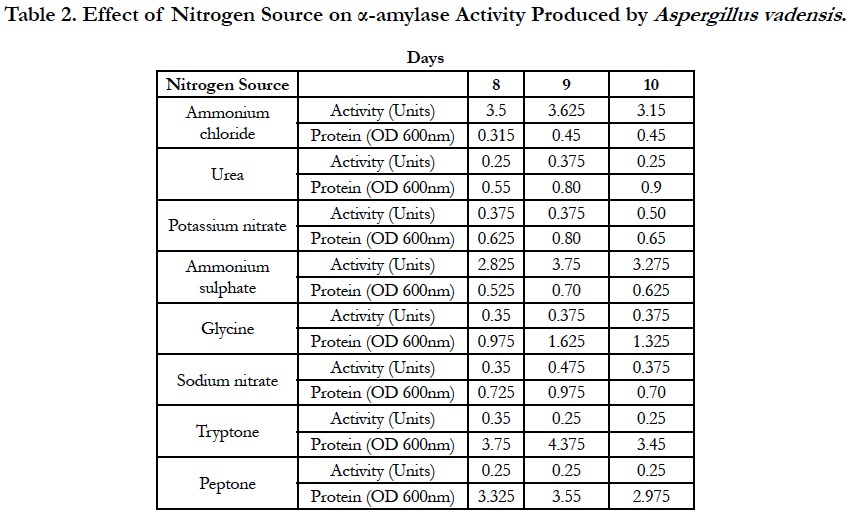
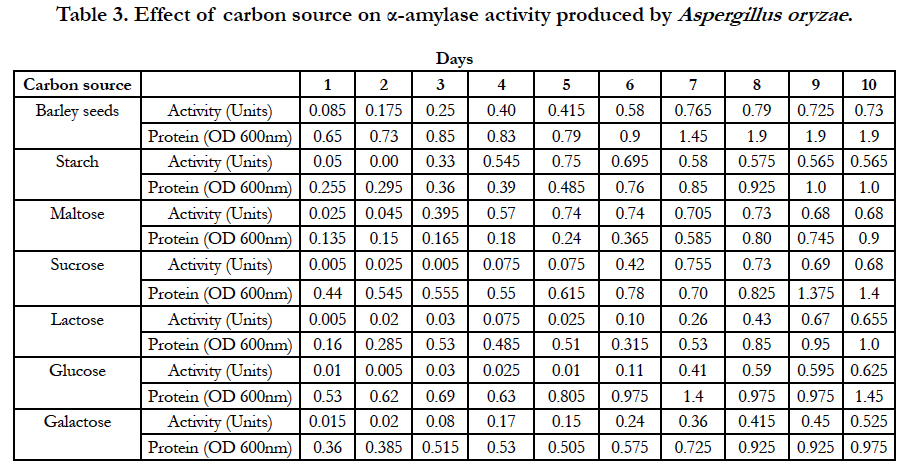
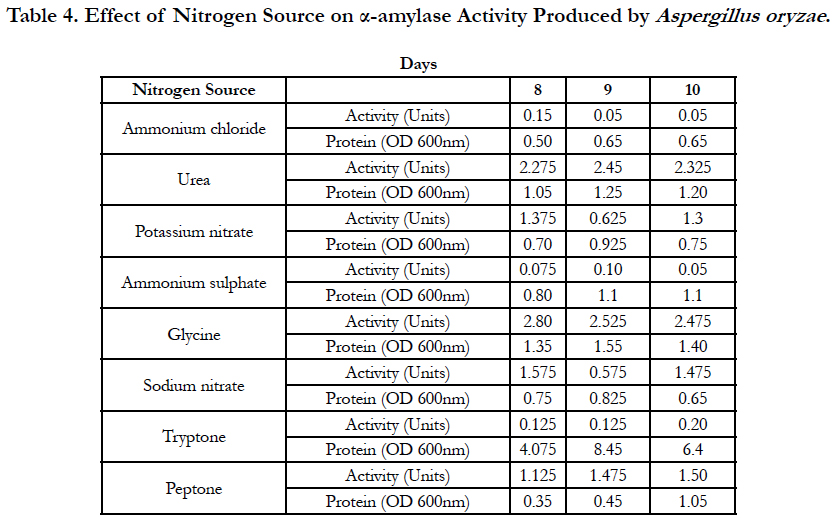
Amylase activities expressed by each isolate in a ten day incubation period at 25°C are presented in Tables 1–4.
Our studies conducted on the fungal strains Aspergillus vadensis revealed that when barley-seed was a carbon source as well as sole nutrient source for fungal growth, the optimum activity of α-amylase produced by this fungus was observed on the 8th day of inoculation of medium and was equal to 0.55 Units. When potassium nitrate was used as a nitrogen source of fungal growth in our defined medium and starch was used as a carbon source, the optimum activity of α-amylase produced by this isolate was observed on the 7th day of inoculation of medium and was equal to 0.47 Units. With maltose used as a carbon source, the optimum activity of α-amylase was also detected on the 7th day of inoculation of medium and was equal to 0.675 Units. When sucrose was used as a carbon source, the optimum activity of α-amylase was observed on the 8th day of inoculation of medium and was equal to 0.565 Units. With lactose used as a carbon source, the optimum activity of α-amylase was produced on day 7th day of inoculation of medium and was equal to 0.45 Units. When glucose was used as a carbon source of fungal growth, the optimum activity of α-amylase was observed on the 7th day of inoculation of medium and was equal to 0.69 Units. When galactose was used as a carbon source, the optimum activity of α-amylase was observed on the 10th day of inoculation of medium and was equal to 0.055 Units (Table 1).
When the carbon source of fungal growth in the defined medium was starch and the nitrogen source was ammonium chloride, the optimum activity of α-amylase produced by Aspergillus vadensis was observed on the 9th day of inoculation of medium and was equal to 3.625 Units. With urea used as a nitrogen source, the optimum activity of α-amylase produced by Aspergillus vadensis was observed on the 9th day of inoculation of medium and was equal to 0.375 Units. When potassium nitrate was used as a nitrogen source, the optimum activity of α-amylase produced by Aspergillus vadensis was observed on the 8th-9th days of inoculation of medium and was equal to 0.375 Units. With ammonium sulphate used as a nitrogen source of fungal growth, the optimum activity of α-amylase produced by Aspergillus vadensis was observed on the 9th day of inoculation of medium and was equal to 3.75 Units.
With glycine used as a nitrogen source, the optimum activity of α-amylase produced by Aspergillus vadensis was observed on the 9th day of inoculation of medium and was equal to 0.375 Units. When sodium nitrate was used as a nitrogen source, the optimum activity of α-amylase produced by Aspergillus vadensis was observed on the 9th day of inoculation of medium and was equal to 0.475 Units. With tryptone used as a nitrogen source, the optimum activity of α-amylase produced by Aspergillus vadensis was observed on the 8th day of inoculation of medium and was equal to 0.35 Units. However, when peptone was used a as nitrogen source, the optimum activity of α-amylase produced by Aspergillus vadensis was observed on the 8th, 9th and 10th days of inoculation of medium and was equal to 0.25 Units (Table 2).
Further studies conducted on the fungal strain Aspergillus oryzae revealed that when barley-seed was a carbon source as well as sole nutrient source for fungal growth, the optimum activity of α-amylase produced by this fungal strain was observed on the 8th day of inoculation of medium and was equal to 0.79 Units. When potassium nitrate was used as a nitrogen source of fungal growth in the medium and starch was used as a carbon source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 5th day of inoculation of medium and was equal to 0.75 Units. With maltose used as a carbon source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 5th and 6th days of inoculation of medium and was equal to 0.74 Units. When sucrose was used as a carbon source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 7th day of inoculation of medium and was equal to 0.755 Units. With lactose used as a carbon source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 9th day of inoculation of medium and was equal to 0.67 Units. When glucose was used as a carbon source of fungal growth, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 10th day of inoculation of medium and was equal to 0.625 Units. When galactose was used as a carbon source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 10th day of inoculation of medium and was equal to 0.525 Units (Table 3).
When starch was used as a carbon source of fungal growth in the defined medium and ammonium chloride was used as nitrogen source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 8th day of inoculation of medium and was equal to 0.15 Units. With urea used as a nitrogen source, optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 9th day of inoculation of medium and was equal to 2.45 Units. When potassium nitrate was used as a nitrogen source, the optimum activity of α-amylase activity produced by Aspergillus oryzae was observed on the 9th day of inoculation of medium and was equal to 0.625 Units. With ammonium sulphate used as a as nitrogen source of fungal growth, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 9th day of inoculation of medium and was equal to 0.10 Units. With glycine used as a as nitrogen source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 8th day of inoculation of medium and was equal to 2.80 Units. When sodium nitrate was used as a nitrogen source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 8th day of inoculation of medium and was equal to 1.575 Units. With tryptone used as a nitrogen source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 10th day of inoculation of medium and was equal to 0.20 Units. When peptone was used as a nitrogen source, the optimum activity of α-amylase produced by Aspergillus oryzae was observed on the 10th day of inoculation of medium and was equal to 1.50 Units (Table 4).
Discussion
Barley (Hordeum vulgare L.) is a member of the grass family and a major cereal family grown in the temperate regions of the globe [36]. It is also grown in cold regions of the tropics such as Jos, Plateau State in tropical Nigeria, West Africa [37]. It is a rich source of essential nutrients such as protein, dietary fibre, vitamin B6, niacin, dietary minerals such as manganese and phosphorous [38]. Raw barley contains approximately 78% carbohydrate, 1% fat, 10% protein, 10% water [39]. It is used in the production of beer and certain distilled beverages [39]. Studies have shown that barley and barley seeds contain both endogenous α-amylases and α-amylase inhibitors [40]. However, autoclaving the barley and barley seeds at a temperature as high as 121°C for 15 minutes is enough to destroy both the endogenous α-amylase and α-amylase inhibitor [41]. With the destruction of the endogenous α-amylase evidenced with nil α-amylase activity observed at the 1st and 2nd days for barley-seed medium inoculated with Aspergillus vadensis, and vice versa the presence of activity on subsequent days is indicative of production of this enzyme by the inoculating fungal strain. Also α-amylase activity was detected starting 1st day (24 hr after inoculation with isolate) with barley-seed medium inoculated with Aspergillus oryzae.
The carbohydrates in the barley seeds are hydrolysed by the enzyme (α-amylases), presumably yielding maltose, maltotriose and α-dextrins [31, 42]. These products of hydrolysis in increasing concentration may serve nutritive and beneficial values to the consumers. Also, the presence of fats and proteins may stimulate the production of other enzymes such as microbial lipases and proteases depending on the infecting phytopathogen of the natural un-harvested barley.
The results observed for inoculated barley seeds in Tables 1 and 2 by the two fungal strains not to follow a natural pattern for enzyme production, that is, activity rising steadily with days of inoculation, reaching a peak and falling gradually. Enzymes are proteins and their presence in the inoculated medium is evidenced with observed values at optical density 600nm readings using the Lowry et al., [33] method. It is therefore suggested that the products of enzymatic (α-amylase) hydrolysis might probably be inhibiting expression and production of the enzyme at the genetic level or that these α-amylases might be targets of hydrolyses by proteases produced by the fungi.
When potassium nitrate was used as a nitrogen source, maltose and glucose were the best carbon sources for fungal growth and production of the most active α-amylases by the fungal strain Aspergillus vadensis at 25°C. Barley seeds, starch and sucrose seemed the best carbon sources for fungal growth and production of α-amylase by the fungal strain Aspergillus oryzae at this same temperature. Ammonium chloride and ammonium sulphate were the best nitrogen sources for production of α-amylase by the fungal strain Aspergillus vadensis when starch is the carbon source in the medium for fungal growth at 25°C. However, urea, glycine and sodium nitrate were the best nitrogen sources for fungal strain Aspergillus oryzae.
Russia and Ukraine are currently recognized as the world’s second largest producers of barley. Nigeria, West Africa is recently being introduced to the production of this highly adaptive cereal. The industrial production of active α-amylases is encouraged using either of the two fungal isolates in Nigeria and Ukraine.
Acknowledgements
Authors thank the British Mycological Society, Britain, The United Kingdom for Grant Support and the Bill and Melinda Gates Foundation for Global Health Support.
References
- Bordbar AK, Omidiyan K, Hosseinzadeh R (2005) Study on interaction of α-amylase from Bacillus subtilis with acetyl trimethylammonium bromide. Colloids and Surfactants B: Biointerfaces. 40(1): 67-71.
- van der Maarel MJ, van der Veen B, Uitdehaag JC, Leemhuis H, Dijkhuizen L (2002) Properties and applications of starch-converting enzymes of the α-amylase family. J Biotechnol. 94(2): 137-155.
- Satyanarayana T, Rao JL, Ezhilvannan M (2005) α-Amylases. In: Enzyme Technology. Asiatech Publishers Inc., New Delhi, India. 189-220.
- Pandey A, Nigman P, Soccol CR, Soccol VT, Singh D, et al., (2000) Advances in microbial amylase. Biotechnol Appl Biochem. 31(2): 135-152.
- Adejuwon A (2013a) Alpha Amylases from Fungi: Species of Aspergilli and Penicilli. LAP Lambert Academic Publishing, Saarbrucken, Deutschland/ Germany. 85pp.
- Cheng C, Udaka S (1991) Efficient production of Taka-amylase A by Trichoderma viride. Agric Biol Chem. 55(7): 1817-1822.
- Kabayashi Y, Onodera R (1999) Application of molecular biology to rumen microbes: Review. Asian-Australasian J Animal Sci. 12(1): 77-83.
- Shin CS, Kim HJ, Kim MJ, Ju JY (1998) Morphological changes and enhanced pigment production of Monascus when cocultured with Saccharomyces cerevisiae or Aspergillus oryzae. Biotechnol Bioeng. 59(5): 576-581.
- Skovgaard K, Rosendahl S (1998) Comparison of intra- and extracellular isozyme banding patterns of Fusarium oxysporum. Mycological Res. 102(9): 1077-1084.
- Legin E, Ladrat C, Godfroy A, Barbier G, Duchiron F (1997) Thermostable amylolytic enzymes of thermophilic microorganisms from deep-sea hydrothermal vents. Comptes Rendus de l’ Academie des Sciences Series III Sciences de la Vie. 320(11): 893-898.
- Adams P (1997) Growth and amylase production in Sporotrichum thermophile Apinis. Biotechnol Appl Biochem. 26(3): 169-170.
- Hayashi K, Nozaki H (1999) Isolation and characterization of gibbestatin B, an inhibitor of gibberelin-induced expression of alpha-amylase and gibbestatin C from Streptomycetaes. J Antibiot. 52(10): 917-920.
- Pinn EH, Rogerson A, Atkinson RJA (1997) Microbial flora associated with the digestive system of Upogebia stellata (Crustacea: Decapoda: Thalassinidea). J Marine Biological Association of the United Kingdom. 77(4): 1083-1096.
- Kato M (1998) Trehalose production with a new enzymatic system from Sulfolobus sulfataricus. J Molec Cataly B Enzymatic. 6(3): 223-233.
- Legin E, Copinet A, Duchiron F (1998) Production of thermostable amylolytic enzymes by Thermococcus hydrothermalis. Biotechnol Lett. 20(4): 363-367.
- Ogawa T, Yoshinori I, Yotome C (1990) Inactivation of DNA polymerase 1, alpha-amylase and protease by basic dyes and inhibition of microbial growth. Bull Environ Contam Toxicol. 45(1): 107-112.
- Douglas DJ, Collings BA, Numao S, Nesatyy VJ (2001) Detection of noncovalent complex between alpha-amylase and its microbial inhibitor tendamistat by electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 15(2): 89-96.
- Kichakova NA, Pavlova IN, Zakharova IY (1998) Purification and identification of amylolytic enzymes from Bacillus licheniformis. Appl biochem microbiol. 34(5): 503-507.
- Purohit JS, Dutta JR, Nanda R, Banerjee R (2006) Strain improvement for tannaseproduction from co-culture of Aspergillus foetidus and Rhizopus oryzae. Bioresource Technol. 97(6): 795-801.
- Moller K, Sharif MZ, Olsson L (2004) Production of fungal alpha-amylase by Saccharomyces kluyveri in glucose-limited cultivations. J Biotechnol. 111(3): 311-318.
- Chen DC, Chen SY, Gee MF, Pan JT (1999) A variant of Saccharomyces cerevisiae pep4 strain with improved oligotrophic proliferation, cell survival and heterologous secretion of alpha-amylase. Appl Microbiology Biotechnol. 51(2): 185-192.
- Davison SP, Santangelo JD, Reid SJ, Woods DR (1995) A Clostridium acetobutylicum regulator gene (regA) affecting amylase production in Bacillus subtilis. Microbiology. 141(4): 989-996.
- Liu G, Zhang Y, Xing M (2006) Dual promoters enhance heterologous enzyme production from bacterial phage based recombinant Bacillus subtilis. Sheng Wu Gong Cheng Xue Bao. 22(2): 191-197.
- Long CM, Virolle MJ, Chang SY, Chang S, Bibb MJ (1987) Alpha-amylase gene of Streptomyces limosus: nucleotide sequence, expression motifs and amino acid sequence homology to mammalian and invertebrate alpha-amylases. J Bacteriol. 169(12): 5745-5754.
- Simpson CL, Russell RRB (1998) Intracellular α-amylase of Streptococcus mutans. J Bacteriol. 180(17): 4711-4717.
- Minetoki T, Kumagai C, Gomi K, Kitamoto K (1998) Improvement of promotor activity by the introduction of multiple copies of the conserved region III sequence, involved in the efficient expression of Aspergillus oryzae amylase-encoding genes. Appl Microbiol Biotechnol. 50(4): 459-467.
- Murygina VP (1978) Natural variability of Bacillus diastaticus, a thremophilic bacterium that forms amylase. Mikrobiologiia. 47(1): 101-106.
- Cannon PF, Kirk PM (2007) Fungal Families of the World. CAB Intl Publi, Wallingford, Oxfordshire. 456.
- Olutiola PO, Ayres PG (1973) Utilization of carbohydrates by Rhynchosporium secalis. I. Growth and sporulation on glucose, galactose and galacturonic acid. Physiologia Plantarum. 29(1): 92-96.
- Olutiola PO, Famurewa O, Sonntag HG (1991) An Introduction to General Microbiology: A Practical Approach. Heidelberger Verlagsanstalt und Druckerei GmbH, Heidelberg, Federal Republic of Germany. 267.
- Adejuwon AO (2011) Nutritional factors affecting the production of α-amylase by Lasiodiplodia theobromae Pat. Biotechnol Bioinf Bioeng. 1(1): 131-135.
- Olutiola PO, Nwaogwugwu RI (1982) Growth, sporulation and production of maltase and proteolytic enzymes in Aspergillus aculeatus. Transactions of the British Mycological Society. 78(1): 105-113.
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the folin phenol reagent. J Biol Chem. 193(1): 265-275.
- Adejuwon AO (2013b) Studies on α-amylases produced by selected fungi associated with deterioration of bread. Ph.D. Thesis, Obafemi Awolowo University, Ile-Ife, Nigeria. 150.
- Pfueller SL, Elliott WH (1969) The extracellular α-amylase of Bacillus stearothemophilus. J Biol Chem. 244(1): 48-54.
- The Australian Government Office of the Gene Technology Regulator (2008) The Biology of Hordeum vulgare L. (Barley). The Department of Health and Ageing, Office of the Gene Technology Regulator. 41.
- Porter G, Phillips-Howard K (1994) Outgrower barley cultivation on the Jos, Plateau, Nigeria: A case study of Agricultural innovation. Geographical J. 160(3): 319-327.
- Aghamirzael M, Heydari-Dalfard A, Karami F, Fathl M (2013) Pseudo-cereals as functional ingredient: Effects of bread nutritional and physiological properties – Review. Intl J Agricult Crop Sci. 5(14): 1574-1580.
- Microbiotic World (2016) Nutritional value, benefits and uses of grain products (gluten-free).
- Vallee F, Kadziola A, Bourne Y, Juy M, Rodenberg KW, et al., (1998) Barley α-amylase bound to its endogenous protein inhibitor BAS1: Crystal structure of the complex at 1.9 a resolution. Structure. 6(5): 649-659.
- Doelle HW, Rockem JS, Berovic M (2009) Biotechnology-Volume V: Fundamentals in Biotechnology. EOLSS Publi. 386.
- Aiyer PV (2005) Amylases and their applications. Afr J Biotechnol. 4(13): 1525-1529.