Male Infertility and Single-Nucleotide Polymorphisms in the Testis-Specific Succinyl CoA: 3-Oxoacid CoA Transferase (SCOT-T/OXCT2) gene in a Japanese Cohort
Tanaka H1*, Miyagawa Y2, Tsujimura A3, Nishimune Y4
1 Faculty of Pharmaceutical Sciences, Nagasaki International University, Nagasaki, Japan.
2 Department of Urology, Graduate School of Medicine, Osaka University, Suita, Osaka, Japan.
3 Department of Urology, Juntendo University Urayasu Hospital, Chiba, Japan.
4 Department of Science for Laboratory Animal Experimentation, Research Institute for Microbial Diseases, Osaka University, Suita, Osaka, Japan.
*Corresponding Author
Hiromitsu Tanaka,
Faculty of Pharmaceutical Sciences, Nagasaki International University,
2825-7 Sasebo, Nagasaki, 859-3298, Japan.
Tel: +81 956-205651
E-mail: h-tanaka@niu.ac.jp
Received: July 19, 2018; Accepted: July 30, 2018; Published: August 02, 2018
Citation: Tanaka H, Miyagawa Y, Tsujimura A, Nishimune Y. Male Infertility and Single-Nucleotide Polymorphisms in the Testis-Specific Succinyl CoA: 3-Oxoacid CoA Transferase (SCOT-T/OXCT2) gene in a Japanese Cohort. Int J Reprod Fertil Sex Health. 2018;S1:02:001:1-6. doi: dx.doi.org/10.19070/2377-1887-SI0201001
Copyright: Tanaka H© 2018. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use,distribution and reproduction in any medium, provided the original author and source are credited.
Annotation: This paper described details of the established patents (WO03/068969 A1) with additional data.
Abstract
Some cases of male infertility show an association with various genetic factors. However, putative causative genes remain inconclusive in the majority of cases and do not explain all types of male infertility. Succinyl CoA: 3-oxoacid CoA transferase (SCOT; EC 2.8.3.5) is a key enzyme for the metabolism of ketone bodies, and its testicular isotype (SCOT-T/OXCT2) is localized exclusively in the cytoplasm of spermatids and the midpiece of ejaculated sperm where mitochondria exist. SCOT-T has been suggested to play important roles in spermiogenesis and sperm function. This study compared the genomic DNA sequence of human SCOT-T in sterile males and proven-fertile control volunteers. We found four single nucleotide polymorphisms (SNPs): one in the 3′ noncoding region, and three in the coding region causing predicted amino acid substitutions. The prevalence of homozygous SNPs causing amino acid changes was significantly higher in infertile patients than in fertile controls. This study suggests that these SNPs have detrimental effects on SCOT-T enzyme activity, which influences male infertility.
2.Abbreviations
3.Introduction
4.Materials and Methods
4.1 Participants
4.2 Identification of SNPs in SCOT-T genomic DNA by direct sequencing of PCR-amplified DNA
4.3 Statistical analysis
5.Results
6.Discussion
7.Acknowledgments
8.References
Keywords
Genome; Ketone; Mutation; Metabolism; Sperm.
Abbreviations
SNPs: Single Nucleotide Polymorphisms; OXCT: Oxoacid CoA-Transferase; PCR: Polymerase Chain Reaction; SCOT: Succinyl-CoA Transferase.
Introduction
Approximately 20% of couples experience some form of infertility problems, of which about half may be attributed to male factors [1, 2]. Causes of impaired male fertility include endocrine disorders, genetic factors, chromosomal abnormalities, environmental factors, and malformations such as cryptorchidism and varicocele [3-6]. However, the underlying causes remain unclear in the majority of male infertility cases involving inadequate spermatogenesis or sperm defects.
Succinyl CoA: 3-oxoacid CoA transferase (OXCT/SCOT) is a key enzyme for energy metabolism of ketone bodies exported to peripheral tissues expressed in the liver [7]. SCOT is localized in the mitochondria, and catalyzes the formation of acetoacetyl CoA by transferring a CoA moiety from succinyl CoA to acetoacetate and raises two acetyl CoA molecules capable of entering the tricarboxylic acid cycle [8]. SCOT cDNAs have been cloned from pigs and human hearts [9, 10].
Previously, germ cell-specific SCOT-T1 and 2 were cloned from a subtracted cDNA library of mouse testis [11, 12]. Mouse SCOT-T was shown to be specifically expressed in late spermatids and localized in mitochondria of sperm by northern blot, Western blot, and immunohistochemical analyses. The nucleotide (nt) sequence of mouse SCOT-T was more than 62% identical and the deduced amino acid (aa) sequence was more than 67% identical to SCOT in pigs and humans [11, 12]. Residues 1-39 at the NH2 terminus of SCOT-T conserved a putative mitochondria targeting signal sequence and a glutamate residue (aa residue: 341) corresponding to glutamate 344, known to be conserved in all CoA transferases in SCOT [14]. Mouse SCOT-T proteins are germ cell-specific isoforms of Scot that exist in the mitochondria of haploid spermatids and sperm where they play specific roles in spermatogenesis and energy metabolism.
We cloned and characterized the human ortholog of mouse SCOT-T. SCOT-T is an intronless gene, like in mouse, and was specifically expressed in the testis [13]. SCOT-T mRNA and the deduced aa sequence of human SCOT-T showed 75.4% and 75.8% identity with mouse SCOT-T, respectively. The mitochondria targeting signal sequence, a glutamate residue, and the hydrophilic bridge are also conserved [13, 14].
Several investigators have reported the importance of mitochondrial enzymes for energy metabolism in sperm motility and function [15-17]. The specific presence of SCOT-T may indicate the existence of a novel metabolic system that utilizes ketone bodies as an energy source for sperm motility [18]. In addition, specific expression in haploid spermatids suggests specialized roles of SCOT-T in spermiogenesis [19]. Mutations or single nt polymorphisms (SNPs) that induce the loss of SCOT-T enzyme activity might be expected to impair spermatogenesis and sperm activity, which may affect fertilization ability in human males.
The intronless genome structure allows the study of genetic changes by direct sequencing of polymerase chain reaction (PCR)-amplified DNA of infertile male patients. The present study assessed the prevalence of the SCOT-T SNPs in male patients seeking fertility evaluation. DNA samples from 516 males (261 proven-fertile volunteers and 255 infertile patients) were analyzed. We identified three SNPs in the SCOT-T genomic DNA, which cause predicted aa substitutions. The prevalence of these SNPs was significantly higher in some of the infertile male patients than in the fertile control population, suggesting a causative role in male infertility.
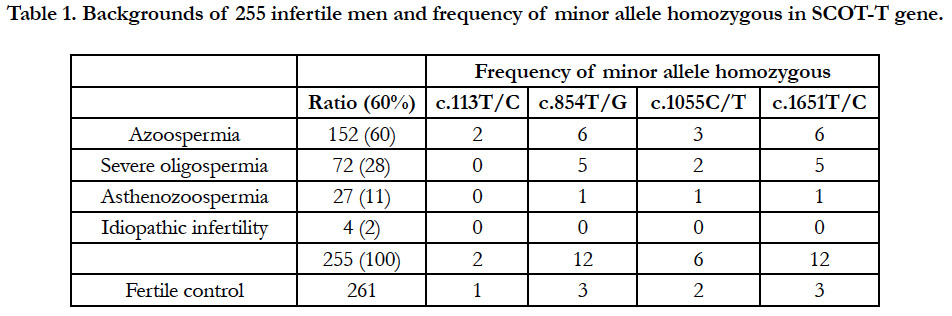
A total of 255 infertile males were divided into subgroups according to the degree of defective spermatogenesis. Accordingly, 152 (60%) had non-obstructive azoospermia, 72 (28%) had severe oligozoospermia (0.1 to 3 × 106 cells/mL), 27 (11%) had low sperm motility, and 4 (2%) had idiopathic infertility with no abnormal sperm morphology and motility (Table 1). The control group was composed of 261 proven-fertile males who had impregnated female patients from the maternity clinic. DNA samples were extracted from the blood leukocytes of infertile and proven-fertile males.
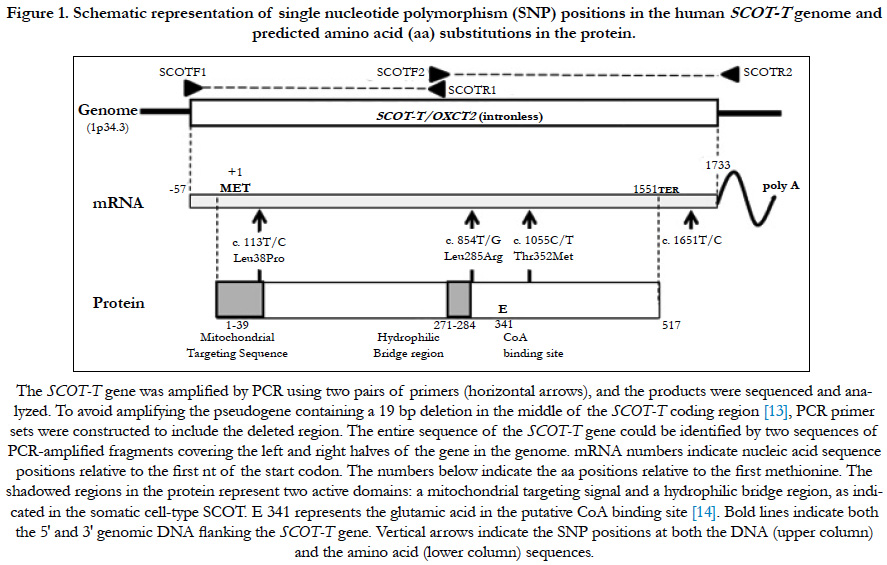
The human genome contains a SCOT-T pseudogene that contains a 19 bp deletion (18 nts from 745-762 and one nt at 778, separated by 15 nts) in the middle of the coding region [13]. Therefore, two PCR primer sets were constructed to include the deleted region, thereby avoiding amplification of the pseudogene. Primer pairs SCOTF1 (TGCTCTGTGACGCGCGGCCCGAGGC) and SCOTR1 (CCTCCACGATCTCTTCCACCTCCACC) were used to amplify the 5′ half of the gene, which included 25 nts upstream of the putative transcription start site through 745. Primer pairs SCOTF2 (CGGTGGAGGTGGAAGAGATCGTGG) and SCOTR2 (TCCATTCCTCACCACTGCAGACCTG) were used to amplify the 3′ half from 744 to 25 nts on the downstream side of the putative transcription stop site of the SCOT-T gene (Figure 1). The entire SCOT-T DNA sequence (except 35 nts in the middle portion around the inner primer sequences: 730-764) could be identified by two sequences of PCR-amplified fragments covering the left and right halves of the SCOT-T gene (Figure 1). PCR reactions were performed using the manufacturer’s recommended reaction buffer (50 μL) containing 0.01 μg human genomic DNA, 0.2 mM of each primer, 2.5 mM each of dGTP, dATP, dCTP, and dTTP, and EX Taq polymerase (Takara, Shiga, Japan). Cycling conditions included 96°C for 2 min, followed by 35 cycles of denaturation at 96°C for 45 s, annealing at 71°C for 45 s, and extension at 72°C for 60 s. PCR-amplified fragments were purified using AMPure (Agencourt Bioscience Corporation, Beverly, MA, USA) and sequenced using the same PCR primers and the Big-Dye Terminator v 3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). The reaction products were purified using BigDye XTerminator and analyzed using an ABI PRISM 3730xl Genetic Analyzer (Applied Biosystems). Screening for sequence variation was performed using SeqScape software (Applied Biosystems).
Figure 1. Schematic representation of single nucleotide polymorphism (SNP) positions in the human SCOT-T genome and predicted amino acid (aa) substitutions in the protein.
The χ2-test was used to compare genotype distributions between infertile subjects and proven-fertile controls. P values < 0.05 were considered to indicate statistical significance.
Results
To compare the genomic DNA sequence of the SCOT-T gene in various infertile patients and fertile controls, we engineered specific primers that precluded amplification of the pseudogene (OXCT2P1) [13]. To amplify the SCOT-T gene, PCR primers were designed at both ends and in the middle portion of the gene that contained 19 bp genomic DNA missing from the non-functional pseudogene (Figure 1) [13]. These selective primers ensured that only the genomic DNA gene was amplified and not the pseudogene. Using PCR and direct sequencing of the amplified DNA, the prevalence of the SCOT-T SNPs was assessed in samples from male patients seeking fertility evaluation and from provenfertile controls.
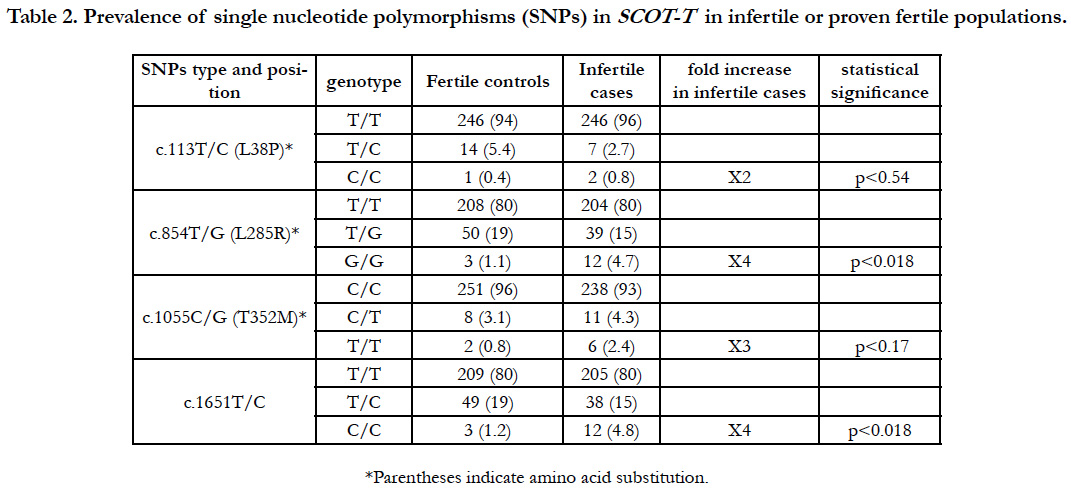
We found four SNPs: one in the 3′ noncoding region, and three in the coding region that caused predicted aa changes at positions 38, 285, and 352 aa (Figure 1) in a total of 516 males (261 fertile volunteer controls and 255 infertile cases) (Table 1). Clinical features of infertile patients investigated in this study and those with these SNPs are listed in Table 1.
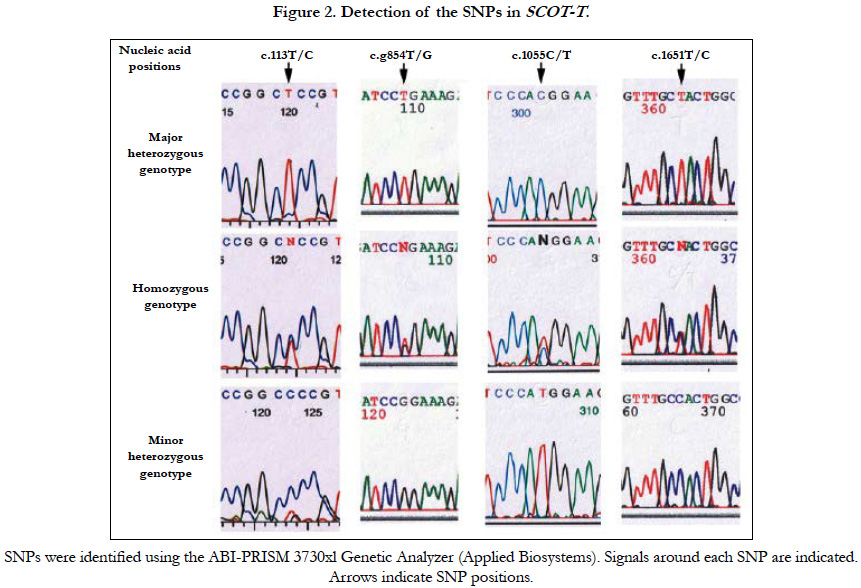
A polymorphism at c.113T/C, which corresponded to aa 38 (L38P) within the consensus mitochondrial targeting sequence domain, showed that 94% (n=246) of controls and 96% (n=246) of infertile cases were homozygous for the major allele (T/T) encoding leucine; 5.4% (n=14) of controls and 2.7% (n=7) of cases were heterozygous (T/C); and 0.4% (n=1) of controls and 0.8% (n=2) of infertile cases were homozygous for the minor allele (C/C), which caused an aa change to proline (Table 2 and Figure 2).
Table 2. Prevalence of single nucleotide polymorphisms (SNPs) in SCOT-T in infertile or proven fertile populations.
The c.854T/G SNP at aa 285 (L285R) showed that 80% (n=208) of controls and 80% (n=204) of infertile cases were homozygous for the major allele that encoded leucine (T/T); 19% (n=50) of controls and 15% (n=39) of cases were heterozygous (T/G); and 1.1% (n=3) of controls and 4.7% (n=12) of infertile cases were homozygous for the minor allele (G/G), causing an aa change to arginine (G/G).
The c.1055C/T SNP at aa 352 (T352M) showed that 96% (n=251) of controls and 93% (n=238) of infertile cases were homozygous for the major allele (C/C) which encodes threonine (C/C); 3.1% (n=8) of controls and 4.3% (n=11) of infertile cases were heterozygous (C/T); and 0.8% (n=2) of controls and 2.4% (n=6) of infertile cases were homozygous for the minor allele (T/T), causing aa change from threonine to methionine.
The prevalence of homozygous SNPs causing aa changes at three regions was significantly higher by two- to four-fold in the infertile population than the fertile controls using statistical analysis (Table 2). Furthermore, we found a SNP (c.1651T/C) in the 3′ non-coding region at which 80% (n=209) of controls and 80% (n=205) of cases were homozygous for the major allele (T/T), 19% (n=49) of controls and 15% (n=38) of cases were heterozygous (T/C), and 1.2% (n=3) of controls and 4.8% (n=12) were homozygous for the minor allele (C/C). Interestingly, we found that nearly all individuals with the minor homozygous c.1651C/C allele had double SNPs, with all controls (n=3) and all infertile cases but one (n=11) also having the minor G/G allele at c.854T/G. Several individuals were also heterozygous at c.1376G/T, c.1439A/G, c.1480A/G, and c.1597A/G in both the fertile and infertile groups among 516 Japanese males. These results suggest that succinyl CoA transferase activity of SCOT-T may be a prerequisite for male fertility, and SNPs affecting the enzyme activity may cause male infertility.
Discussion
Many genetic factors have been associated with differences in human sperm production, including deletion of Y-chromosomal regions [20], gene mutations in methylene tetrahydrofolate reductase [21] or at the mitochondrial DNA polymerase locus [22], some molecular variants of LH [23] and HLA genes [24]; polymorphisms of cytochrome P450–1A1 [25]; and CAG repeat length in the androgen receptor [26, 27]. Also, there are many other genetic etiologies, such as chromosomal rearrangement, for infertility. However, the cause of many cases of idiopathic male infertility is unknown.
SCOT-T, an intronless gene, and a SCOT-T-like pseudogene [13] are both located in the human genome on chromosome 1 (1p33– 35.4) [28] where many chromosome rearrangements have been reported in infertile males [29]. On the other hand, human somatic cells ubiquitously express SCOT on chromosome 5 (5p13.1) from a gene that spans more than 100 kb and contains 17 exons [10]. The lack of introns in SCOT-T and the presence of the SCOT-T-like pseudogene indicate that SCOT-T formed from a retrotransposon, a class of genes created by reverse transcription of a mature mRNA of an ancestor gene. In this manner, a DNA copy of a gene common to SCOT from somatic cells was inserted into the genome [10, 13]. The SCOT-T-like pseudogene has 5 nt differences as well as a 19 bp deletion relative to SCOTT, whereas both SCOT-T1 and SCOT-T2 appear functional in mice [11, 12]. The 19 kbp DNA fragment containing SCOT-T in chromosome 1p34.1–35.3 (AL033527) was likely translocated to a locus in chromosome 1p33–34.3 (AL033526) and changed to a pseudogene [13, 28].
Ubiquitously expressed in somatic cell types, SCOT is known to play a key role in energy metabolism using ketone bodies. Several groups have reported cases of ketoacidosis due to SCOT deficiency caused by a nonsense or missense mutation in the gene [10, 30, 31]. Clinical features and disease severities differ among patients with mutations in SCOT, which may depend in part on the enzyme activity of SCOT. As SCOT-T is also a mitochondrial enzyme for energy metabolism of ketone bodies exclusively expressed in haploid spermatids and sperm, it is possible that loss of SCOT-T activity due to SNPs in SCOT-T may cause some impairment of spermiogenesis and/or sperm function and result in male infertility.
Several consensus regions in SCOT have been proposed in somatic cell types in various animals and organisms, such as a mitochondrial targeting sequence, hydrophilic bridge regions related to the dimerization of SCOT enzyme, and the SENG where the CoA-binding glutamate residue exists [13]. Immunohistochemical analysis has demonstrated that SCOT-T localizes to the cytoplasm of spermatids and also to the midpiece of sperm where mitochondria are found, and a 39-aa N-terminal mitochondrial targeting signal is conserved. Human SCOT-T contains other consensus regions of SCOT [13]. The SNP (c.854T/G (L285R)) observed exclusively in infertile males is localized near the hydrophilic bridge region. The resulting ‘aa’ change may affect the enzyme activity by some inhibitory effect of dimerization of SCOTT. Furthermore, SNPs in the noncoding region (c.1651T/C) may affect mRNA stability and/or translation for enzyme production, which may cause the impairment of SCOT-T. SNPs in the noncoding region (c.1651T/C) and in c.854T/G (L285R) were observed in 15 patients. Two SNPs were linked in all but one infertile patient. These two SNPs may influence enzyme activity, including effective translation and intracellular localization.
Although this study investigated only a limited number of cases, the results strongly suggest that SNPs causing impairment of SCOT-T enzyme might affect male fertility. The clinical features and SNPs of the infertile patients investigated in this study are listed in Table 1. The majority of the homozygous SNPs at L285R occurred in cases with azoospermia or severe oligozoospermia, some of which were also accompanied by defective mobility (asthenozoospermia). Such conditions might result from disruption of the two developmental functions of SCOT-T: the morphogenesis of haploid spermatids (spermiogenesis) and the function of sperm. It would be interesting to investigate the enzyme activity of SCOT-T in the sperm of patients with oligozoospermia and who have the identified homozygous SNPs; however, collection of the clinical samples would be difficult.
At this time, it is not possible to completely elucidate the relationship between SNPs and clinical features while simultaneously anonymizing DNA samples. Further screening of a greater number of human DNA samples from infertile and proven-fertile males is required to confirm the clinical phenotype caused by SCOT-T SNPs. Such confirmation may be useful for developing a novel method for diagnosing and treating specific types of male infertility.
In this study, significant differences in the frequencies of the genotypes associated with SCOT-T genetic polymorphisms were identified in infertile subjects (P>0.05). In conclusion, the negative effects of certain SNPs on SCOT-T enzyme activity could be one of the causes of male infertility. These results will significantly contribute to future large-scale studies on the genetic background of infertility in Japanese males and to enzymatic analyses of the role of SCOT-T.
Acknowledgments
We thank Ms. H. Nishimuna for her technical assistance with sequence analysis of human DNA samples.
References
- National Institute of Population of Social Security Research. 14th birth trend survey. Tokyo, Japan; 2010.
- Schlegel PN. Evaluation of male infertility. Minerva Ginecol. 2009 Aug;61(4):261-83. PubMed PMID: 19745794.
- Nishimune Y, Tanaka H. Infertility caused by polymorphisms or mutations in spermatogenesis - specific genes. J Androl. 2006 May-Jun;27(3):326-34. PubMed PMID: 16474012.
- Krausz C, Giachini C. Genetic risk factors in male infertility. Arch Androl. 2007 May-Jun;53(3):125-33. PubMed PMID: 17612870.
- O’Flynn O'Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: a review. Fertil Steril. 2010 Jan;93(1):1-12. doi: 10.1016/j.fertnstert.2009.10.045. PubMed PMID: 20103481.
- Tainaka H, Takahashi H, Umezawa M, Tanaka H, Nishimune Y, Oshio S, et al. Evaluation of the testicular toxicity of prenatal exposure to bisphenol A based on microarray analysis combined with MeSH annotation. J Toxicol Sci. 2012;37(3):539-48. PubMed PMID: 22687993.
- Williamson DH, Bates MW, Page MA, Krebs HA. Activities of enzymes involved in acetoacetate utilization in adult mammalian tissues. Biochem J. 1971 Jan;121(1):41-7. PubMed PMID: 5165621.
- Tildon JT, Cornblath M. Succinyl-CoA: 3-ketoacid CoA-transferase deficiency. A cause for ketoacidosis in infancy. J Clin Invest. 1972 Mar;51(3):493-8. PubMed PMID: 4258782.
- Lin TW, Bridger WA. Sequence of a cDNA clone encoding pig heart mitochondrial CoA transferase. J Biol Chem. 1992 Jan 15;267(2):975-8. Pub- Med PMID: 1730685.
- Kassovska-Bratinova S, Fukao T, Song XQ, Duncan AM, Chen HS, Robert MF, et al. Succinyl CoA: 3-oxoacid CoA transferase (SCOT): human cDNA cloning, human chromosomal mapping to 5p13, and mutation detection in a SCOT-deficient patient. Am J Hum Genet. 1996 Sep;59(3):519-28. PubMed PMID: 8751852.
- Koga M, Tanaka H, Yomogida K, Nozaki M, Tsuchida J, Ohta H, et al. Isolation and characterization of a haploid germ cell-specific novel complementary deoxyribonucleic acid; testis-specific homologue of succinyl CoA: 3-Oxo acid CoA transferase. Biol Reprod. 2000 Dec;63(6):1601-9. PubMed PMID: 11090426.
- Tanaka H, Koga M, Yomogida K, Iguchi N, Nozaki M, Onishi M. Isolation and characterization of haploid germ cell-specific succinyl CoA: 3-oxo Acid CoA Transferase (SCOT-T1 and SCOT-T2). In Andrology in the 21st Century: Proceeding of VIIth International Congress of Andrology; 2001. p. 157-161.
- Tanaka H, Kohroki J, Iguchi N, Onishi M, Nishimune Y. Cloning and characterization of a human orthologue of testis-specific succinyl CoA: 3-oxo acid CoA transferase (SCOT-T) cDNA. Mol Hum Reprod. 2002 Jan 1;8(1):16-23.
- Rochet JC, Bridger WA. Identification of glutamate 344 as the catalytic residue in the active site of pig heart CoA transferase. Protein Sci. 1994 Jun;3(6):975-81. PubMed PMID: 7915164.
- Pascual ML, Cebrian-Perez JA, Lopez-Perez MJ, Muino-Blanco T. Shortterm inhibition of the energy metabolism affects motility but not surface properties of sperm cells. Biosci Rep. 1996 Feb;16(1):35-40. PubMed PMID: 8861538.
- Yeung CH, Majumder GC, Rolf C, Behre HM, Cooper TG. The role of phosphocreatine kinase in the motility of human spermatozoa supported by different metabolic substrates. Mol Hum Reprod. 1996 Aug;2(8):591-6. PubMed PMID: 9239671.
- Ruiz-Pesini E, Diez C, Lapeña AC, Pérez-Martos A, Montoya J, Alvarez E, et al. Correlation of sperm motility with mitochondrial enzymatic activities. Clin Chem. 1998 Aug;44(8 Pt 1):1616-20. PubMed PMID: 9702947.
- Tanaka H, Takahashi T, Iguchi N, Kitamura K, Miyagawa Y, Tsujimura A, et al. Ketone bodies could support the motility but not the acrosome reaction of mouse sperm. Int J Androl. 2004 Jun;27(3):172-7. PubMed PMID: 15139973.
- Tanaka H, Iguchi N, Miyagawa Y, Koga M, Kohroki J, Nishimune Y. Differential expression of succinyl CoA transferase (SCOT) genes in somatic and germline cells of the mouse testis. Int J Androl. 2003 Feb;26(1):52-6. PubMed PMID: 12534938.
- de Vries JW, Hoffer MJ, Repping S, Hoovers JM, Leschot NJ, van der Veen F. Reduced copy number of DAZ genes in subfertile and infertile men. Fertil Steril. 2002 Jan;77(1):68-75. PubMed PMID: 11779593.
- Bezold G, Lange M, Peter RU. Homozygous methylenetetrahydrofolate reductase C677T mutation and male infertility. N Engl J Med. 2001 Apr 12;344(15):1172-3. PubMed PMID: 11302150.
- Rovio AT, Marchington DR, Donat S, Schuppe HC, Abel J, Fritsche E, et al. Mutations at the mitochondrial DNA polymerase (POLG) locus associated with male infertility. Nat Genet. 2001 Nov;29(3):261-2. PubMed PMID: 11687794.
- Ramanujam LN, Liao WX, Roy AC, Ng SC. Association of molecular variants of luteinizing hormone with male infertility. Hum Reprod. 2000 Apr;15(4):925-8. PubMed PMID: 10739843.
- Van der Ven K, Fimmers R, Engels G, Van der Ven H, Krebs D. Evidence for major histocompatibility complex-mediated effects on spermatogenesis inhumans. Hum Reprod. 2000 Jan;15(1):189-96. PubMed PMID: 10611211.
- Fritsche E, Schuppe HC, Döhr O, Ruzicka T, Gleichmann E, Abel J. Increased frequencies of cytochrome P4501A1 polymorphisms in infertile men. Andrologia. 1998 May-Jun;30(3):125-8. PubMed PMID: 9635090.
- Dowsing AT, Yong EL, Clark M, McLachlan RI, de Kretser DM, Trounson AO. Linkage between male infertility and trinucleotide repeat expansion in the androgen-receptor gene. Lancet. 1999 Aug 21;354(9179):640-3. PubMed PMID: 10466666.
- Rajpert-De Meyts E, Leffers H, Petersen JH, Andersen AG, Carlsen E, Jørgensen N, et al. CAG repeat length in androgen-receptor gene and reproductive variables in fertile and infertile men. Lancet. 2002 Jan 5;359(9300):44-6. PubMed PMID: 11809188.
- Onishi M, Yasunaga T, Tanaka H, Nishimune Y, Nozaki M. Gene structure and evolution of testicular haploid germ cell-specific genes, Oxct2a and Oxct2b. Genomics. 2004 Apr;83(4):647-57. PubMed PMID: 15028287.
- Olesen C, Hansen C, Bendsen E, Byskov AG, Schwinger E, Lopez-Pajares I, et al. Identification of human candidate genes for male infertility by digital differential display. Mol Hum Reprod. 2001 Jan;7(1):11-20. PubMed PMID: 11134355.
- Fukao T, Mitchell GA, Song XQ, Nakamura H, Kassovska-Bratinova S, Orii KE, et al. Succinyl-CoA: 3-ketoacid CoA transferase (SCOT): cloning of the human SCOT gene, tertiary structural modeling of the human SCOT monomer, and characterization of three pathogenic mutations. Genomics. 2000 Sep 1;68(2):144-51. PubMed PMID: 10964512.
- Song XQ, Fukao T, Watanabe H, Shintaku H, Hirayama K, Kassovska‐ Bratinova S, et al. Succinyl‐CoA: 3‐ketoacid CoA transferase (SCOT) deficiency: Two pathogenic mutations, V133E and C456F, in Japanese siblings. Hum Mutat. 1998;12(2):83-8. PubMed PMID: 9671268.