Micro-RNA in Health and Disease
Demirtas D1*, Demirtas AO2, Biskin A3, Demirtas M4
1 Department of Internal Medicine, Adana City Hospital, Health Science University, Turkey.
2 Department of Cardiology, Adana City Hospital, Health Science University, Turkey.
3 Department of Genetics, Cukurova University, Turkey.
4 Department of Cardiology, Cukurova University, Turkey.
*Corresponding Author
Derya Demirtas,
Department of Internal Medicine, Adana City Hospital,
Health Science University, Turkey.
E-mail: deryademirtas83@gmail.com
Received: June 08, 2018; Accepted: July 24, 2018; Published: July 31, 2018
Citation: Demirtas D, Demirtas AO, Biskin A, Demirtas M. Micro-RNA in Health and Disease. Int J Cardiol Res. 2018;5(3):116-123. doi: http://dx.doi.org/10.19070/2470-4563-1800020
Copyright: Demirtas D© 2018. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
Micro-RNAs (miRNA) are functional, non-protein coding RNA molecules that are approximately 22 nucleotides in length and their transcriptions provided by intron or exon regions of the genome and non-protein coding regions of RNA genes. miRNAs inhibit translation of protein and destroy mRNA. Although approximately 2500 human miRNA types have been identified so far, quite little is known about their biological functions. In some studies, it is shown that miRNA expression levels are important for many biological processes such as cell proliferation, differentiation, apoptosis, organogenesis and metabolism. Recent studies have pointed out that miRNAs have been used as diagnosis and prognosis biomarkers in many human diseases including cancer, cardiovascular disorders, cerebrovascular disorders and metabolic disturbances.
2.Abbreviations
3.Introduction
4.A Historical Overview of miRNA Research
5.The Biogenesis of miRNAs
6.miRNAs in Human Genome
7.miRNAs and Human Diseases
8.miRNA as a Therapeutic Modality
9.miRNAs in Obesity
10.miRNAs in Hyperlipidemia and Atherosclerosis
11.MiRNAs in Cardiovascular Disease
12.Role of miRNA in Diabetes Mellitus
13.Role of miRNA in Cancer
14.miRNA Regulation on Immunity
15.miRNA and Cerebrovascular Disease
16.Conclusions
17.References
Keywords
Noncoding RNA; miRNA; Biogenesis of miRNA.
Abbreviations
miRNA: Micro RNAs; T2DM: Type 2 Diabetes Mellitus; ORFs: Open Reading Frames; MSCs: Mesenchymal Stem Cells; TC: Total Cholesterol; TG: Triglycerides; LDL-C: Low-Density Lipoprotein Cholesterol; LDL: Low- Density Lipoprotein; VCAM-1: Vascular Cell Adhesion Molecule 1; AMI: Acute Myocardial Infarction; cTnI: Cardiac Troponin I; STEMI: ST-Segment Elevation Myocardial Infarction; CK-MB: Creatine Kinase-MB; PD: Parkinson’s Disease; ND: Neurodegenerative Diseases; AD: Alzheimer’s Disease.
Introduction
Micro RNAs (miRNA) are small noncoding, endogenous, single stranded RNAs usually consisting of 18-25 nucleotides that regulate gene expression through repression or degradation of targeted mRNAs at the post transcriptional level [1]. According to a miRBase online database (http://microrna.sanger.ac.uk/, update on June 26, 2014), more than 2500 microRNAs have been found in human tissues [2]. It is estimated that about 30–50% of protein-coding genes are regulated by miRNAs [3]. The expression of miRNA is controlled by different mechanisms including the level of methylation of DNA and histones [4, 5]. Disrupted expression of miRNAs participating in cell process is related to many diseases, such as obesity, hyperlipidemia, atherosclerosis, type 2 diabetes mellitus (T2DM) and cancer through regulation of multiple genes [6].
A Historical Overview of miRNA Research
miRNAs were discovered in 1993 by Lee and colleagues in the nematode Caenorhabditis elegans. In these organisms, the down regulation of LIN-14 protein was found to be essential for the progression from the first larval stage (L1) to L2. Furthermore, the down regulation of LIN-14 was found to be dependent on the transcription of a second gene called lin-4. Interestingly, the transcribed lin-4 was not translated into a biologically active protein. Instead, it gave rise to two small RNAs approximately 21 and 61 nucleotides in length. The longer sequence formed a stem-loop structure and served as a precursor for the shorter RNA [7]. Later this group, along with Wightman et al., found that the smaller RNA had antisense complementarity to multiple sites in the 3′ UTR of lin-14 mRNA. The binding between these complementary regions decreased LIN-14 protein expression without causing any significant change in its mRNA levels. These two studies together brought forth a model wherein base pairing occurred between multiple lin-4 small RNAs to the complementary sites in the 3′ UTR of lin-14 mRNA, thereby causing translational repression of lin-14 and subsequent progression from L1 to L2 during C. elegans development [8]. This novel mode of regulating gene expression was first thought to be a phenomenon exclusive to C. elegans. In 2000, two separate groups discovered that a small RNA, let-7, was essential for the development of a later larval stage to adult in C. Elegans [9, 10]. More importantly, homologs of this gene were subsequently discovered in many other organisms, including humans [11]. The period that followed was marked by a deluge of information wherein multiple laboratories cloned numerous small RNAs from humans, flies, and worms. These RNAs were noncoding, around 19 to 24 nucleotides in length, and derived from a longer precursor with a stem-loop or fold-back structure [1]. Many were found to be evolutionarily conserved across species and exhibited cell-type specificity. The recognition and confirmation of the existence of these small RNAs, now termed mi-RNAs, led to intense research aimed at identifying new members of this family. This resulted in the discovery of multiple miRNAs across different species of plants and animals. A miRNA registry, named miRBase, set up in 2002 serves as the primary online repository for all potential miRNA sequences, annotation, nomenclature, and target prediction information [12, 13]. The current release (miRBase 20) contains 24 521 entries representing hairpin precursor miRNAs that express 30 424 mature miRNA products in 206 species. The biological significance of a vast majority of annotated miRNAs, however, remains unknown and requires functional validation.
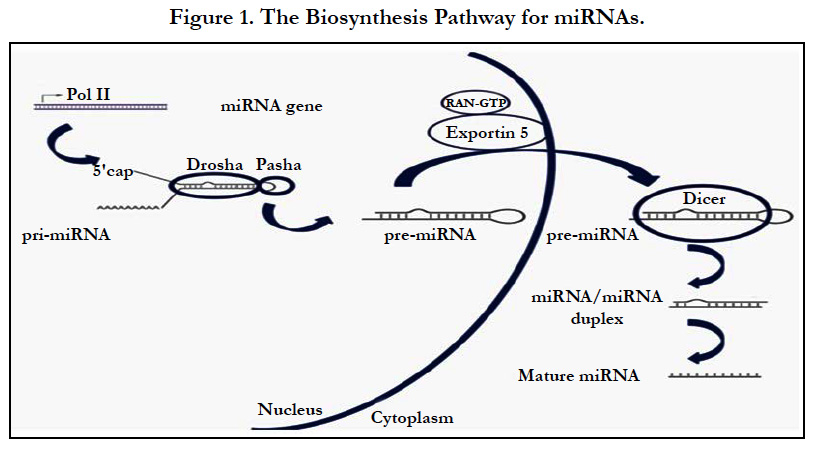
The Biogenesis of miRNAs
The biogenesis of miRNAs (Figure 1) involves multiple steps and specific cellular machinery [14]. Each one of the multiple steps that compose miRNA biosynthesis seems to be remarkably well coordinated. Drosha initiates the processing by specific cropping of the stem-loop precursor in the nucleus [15]. The resulting structure, precursor miRNA (pre-miRNA), seems to be a signature motif for all dsRNAs that are involved in small- RNA pathways. Exportin-5 recognizes this signature motif and exports pre-miRNAs to the cytoplasm through nuclear pores on a GTP–GDP gradient [16, 17]. Following export, pre-miRNA is handed over to another RNase III enzyme, Dicer, that dices the pre-miRNA into a miRNA duplex that is further unwinded giving rise to the mature functional miRNA molecule.
miRNAs in Human Genome
miRNAs regulate many major cellular functions such as development, differentiation, growth, and metabolism and approximately 2500 miRNA genes have been reported to exist in the mammalian genome [18]. One-third of the human genome is estimated to be regulated by miRNAs [19]. The precise mechanism involved in the miRNA transcription is not known but proximity to other genes in the genome and their locations in introns of coding genes, noncoding genes and exons are reported to influence their expression [20]. In the genome, miRNAs are organized in clusters and share the same transcriptional regulatory units and are independently expressed if they have their own promoters [21, 22].
Most recent sequence analyses of the human genome demonstrates that the protein coding genes may be as low as 25,000 [23]. Although the exact number of the protein coding genes in the human genome is not known, the 25,000 figure is at least 3-4 times lower than the figure believed in late 1980's. What these new data reveal is that a large segment of the human genome consists of non-coding protein genes. Further sequence analysis indicates that the Open Reading Frames (ORFs) comprise less than 2%, repetitive sequences around 46% [24, 25] and non-coding parts of protein-coding genes (introns, 5′ and 3′-UTRs) an estimated 25–27% of the 3.2 billion bases in the human genome [26].
miRNAs and Human Diseases
miRNAs have been demonstrated to play a major role in a wide range of developmental processes including metabolism, cell proliferation, apoptosis, developmental timing, and neuronal cell fate [27-29]. Other regulatory roles include neuronal gene expression [30], brain morphogenesis [31], muscle differentiation [32], and stem cell division [33].
MicroRNA’s deficiencies or excesses have been linked to some other clinically important diseases ranging from myocardial infarction to autoimmune diseases [34].
miRNA as a Therapeutic Modality
miRNAs work by highly specific binding to the complementary site on the mRNA target and are considered new therapeutic strategies. Specific oligomers, called antagomirs, have a complementary sequence to a specific miRNA and ultimately compete with the target mRNA to bind to miRNA. This concept has shown promise in cell culture models as well as initials in vivo experiments. For example, Krutzfeldt et al., demonstrated that miRNA antagomirs were successfully delivered in vivo and had higher stability showing target modulation in specific tissues where particular miRNAs were expressed. For example, miR-122 antagomir in the liver showed up-regulation of several genes targeted by miR-122 [35]. Conversely, it is possible to restore miRNAs that are often down-regulated in cancer by external delivery using suitable carrier systems. One of the key issues for this modality is to target the specific sites or tissues of interest. Efficient delivery systems for in vivo delivery of miRNA are highly desirable and several are currently under investigation. There are also emerging studies involving restoration of tumor suppressive miRNAs in tumor by in vivo delivery [36].
miRNAs in Obesity
Obesity is a chronic medical condition resulting from increased fat mass and energy storage in adipose tissue, genetic and environmental predispositions, increased calorie uptake, and decreased physical activity, which leads to an adverse effect on health [37]. Adipose tissue is a major contributor to the pathophysiology of obesity. In recent years, there has been substantial attention paid to the role of miRNAs in regulating adipogenesis and obesity [38]. miRNAs can enhance or suppress adipogenic differentiation of mesenchymal stem cells (MSCs) and mature adipocyte differentiation by regulating transcription factors and signaling pathways related to adipogenesis [39]. These miRNAs play important roles in physiologic and pathophysiological conditions which participate in cell differentiation, proliferation, apoptosis, hematopoiesis, limb morphogenesis, and important metabolic pathways, such as insulin secretion, triglyceride and cholesterol biosynthesis, and oxidative stress [40, 41]. Among these, it is shown that miR-103, miR-107, and miR-143 accelerate fat cell development [42]. miR-935, miR-4772, miR-223, and miR-376b are reporters of diet-induced obesity [43]. Mice lacking miR-378 are resistant to obesity and exhibit enhanced mitochondrial fatty acid metabolism and elevated oxidative capacity in insulin target tissues [44]. miR-221, miR-28, and miR-486 are associated with BMI, percentage fat mass, waist, and regional fat distribution [45].
miRNAs in Hyperlipidemia and Atherosclerosis
Hyperlipidemia, a chronic disorder with high levels of triglycerides (TG, hypertriglyceridemia), total cholesterol (TC, hypercholesterolemia), and low-density lipoprotein cholesterol (LDL-C) and a decreased level of high-density lipoprotein cholesterol (HDLC) [46].
Atherosclerosis is a complex multifactorial disease triggered and maintained by a low-level chronic inflammation of the arterial wall [47]. The onset of atherosclerosis includes a dysfunction of endothelial cells, caused by a variety of external stimuli (e.g. hypertension, reactive oxygen species, or modified low-density lipoprotein (LDL) cholesterol). The damaged endothelium consequently begins to express more adhesive molecules, for example, vascular cell adhesion molecule 1 (VCAM-1), leading to promoted adhesion and infiltration by immune system cells [47]. Hyperlipidemia is a well-accepted risk factor in the development of atherosclerosis [48]. Severe hyperlipidemia promotes the progression of atherosclerosis and its end-points, that is, myocardial infarction or stroke, as shown in patients with familial hypercholesterolemia who suffer a myocardial infarction or stroke at a very early age [49, 50].
Several genes and cytokines have been identified as risk factors for atherosclerosis [51, 52] and recent studies have suggested that miRNAs may play a role in regulating the atherosclerotic process [53, 54]. Expression of 1 to 900 microRNAs potentially associated with atherosclerosis was investigated in different studies. Cipollone et al reported that five miRNAsd miR-100, miR-127, miR-145, miR133a, and miR-133bd were over expressed in symptomatic plaques [55]. Among these five miRNAs, miR-145 and miR133a were further shown to modulate stroke-related proteins in vitro. Li et al demonstrated that the levels of miR-21, miR130a, miR-27b, let-7f, and miR-210 increased significantly in the sclerotic intimal samples compared with normal intimal samples from the same patients with atherosclerotic obliterans [56], whereas miR- 221 and miR-222 decreased significantly. Furthermore, significant increases in miR130a, miR-27b, and miR-210 expression were observed in the serum. Another study reported that the expressions of miR-21, miR-34a, miR-146a, miR-146b-5p, and miR-210 were significantly upregulated in human atherosclerotic arteries versus nonatherosclerotic arteries [57]. Fichtlscherer et al., demonstrated that the circulating levels of miR-126, miR- 17, miR92a, miR-145, and miR-155 were significantly reduced in patients with coronary artery disease compared with healthy controls. In contrast, miR-133a and miR-208a levels tend to be higher in patients with coronary artery disease [58].
MiRNAs in Cardiovascular Disease
In the cardiovascular system, miRNAs control the proliferation and differentiation of stem and progenitor cells, and the function of cardiac myocytes, pacemaker cells, endothelial cells and smooth muscle cells. miRNAs play a crucial role in cardiac development and regeneration. They are involved in cardiovascular pathophysiology and their expression is altered in various cardiovascular diseases [59].
miRNA-22 (miR-22) is one of the most abundant miRNA in the heart. Many studies have demonstrated that miR-22 plays critical roles in myocardial infarction and subsequent cardiac remodeling [60].
miR-208 is a cardiac-enriched miRNA and is found at much higher level during cardiac tissue injury [61]. So There was a good correlation between the changes in plasma miR-208 and cardiac troponin I (cTnI), a classical biomarker of acute myocardial infarction (AMI) [62].
miR-208a was absolutely identified to be expressed in the human heart. miR-208a was undetectable in plasma of healthy individuals or non-AMI patients (such as patients with acute kidney injury, chronic renal failure, stroke, or trauma), but it was easily detected in 90.9 % AMI patients and in 100 % AMI patients within 4 h of the onset of chest pain while cTnI was not yet affected, suggesting that miRNAs may leak into the bloodstream at an earlier stage of myocardial injury (the biological peak of troponins is ~18 h after AMI). It is reasonable to speculate that circulating miR-208a has advantages over cTnI (or cTnT) in distinguishing pure AMI from patients with renal disease, stroke, and trauma [63].
In case of acute ST-segment elevation myocardial infarction (STEMI) patients, the levels of plasma miR-208b increased 3,000- fold compared to healthy controls within 12 h after infarction. Peak values of miR-208b were well correlated with peak cTnI and the ejection fraction, indicating a possible role for circulating miR- 208b as a biomarker in diagnosis of STEMI or in prediction of long-term complications [64].
Plasma level of miR-499 was obviously increased in all patients with AMI but it was below the limit of detection for acute coronary syndromes, congestive heart failure, and for all healthy controls. These results indicated that circulating miR-499 might be a useful biomarker for the diagnosis of AMI [65].
In a rat AMI model, plasma level of miR-133 a was increased at 1-3 h, peaked at 3-12 h, and decreased at 12-24 h after coronary artery ligation. In AMI patients, miR-133 plasma level was substantially higher compared with healthy controls. A positive correlation was also reported between the elevated plasma miR-133 and cTNI. These results provided evidence that miR-133 is a powerful biomarker for the diagnosis of AMI [63].
The level of circulating miR-1 was increased nearly 100-fold in AMI patients compared with healthy subjects and showed a positive correlation with serum creatine kinase-MB (CK-MB) level. In another separate clinical study, plasma miR-1 level was significantly higher in AMI patients compared with non-AMI subjects and the level was returned to normal on discharge following medication. Interestingly, the increased plasma miR-1 was not associated with age, gender, blood pressure, diabetes mellitus, or other established biomarkers for AMI including cTnI and CK-MB. Moreover, in a necrosis model of cultured cardiac cells, miR-1 was found to be released into the culture medium and was stable at least for 24 h. Collectively, these results strongly support that circulating miR-1 might be a novel sensitive biomarker for AMI diagnosis [66].
MiR-214, which increases in mouse and human tissue after myocardial infarction, exerts a protective effect during ischemia/ reperfusion. Deletion of miR-214 increases injury and mortality following myocardial infarction [59].
Cai et al., demonstrated that miR-765 is overexpressed in failing hearts and is involved in contractile regulation [67]. Potous et al., revealed the role of miR-126 in right ventricle (RV) failure associated with pulmonary arterial hypertension (PAH). Due to the methylation process, miR-126 is downregulated in RV failure in PAH patients, causing decreased capillary density. Administration of miR-126 mimics ameliorates microvascular density, improves RV function and diminishes fibrosis, whereas antagomiR-mediated miR-126 downregulation exacerbates RV failure [68]. Circulating miRNA-423-5p is also correlated with NTpro BNP. Also, meticulous screening of 186 miRNAs uncovered four main miRNAs (miR-423-5p, miR-22, miR-320a, and miR- 92b) significantly increased in the serum of HF patients. With the detection of the four miRNAs, a sensitive and specific score could be defined for assessing HF patients. The miRNA-score was closely related to several important prognostic parameters, including increased serum BNP, widening QRS, and dilatation of left ventricle and atrium [69, 70].
Role of miRNA in Diabetes Mellitus
Diabetes mellitus (DM) is a group of chronic metabolic diseases characterized by insulin deficiency and/or insulin resistance that leads to elevated blood glucose levels as well as abnormal fat and protein metabolism [68]. The number of people with diabetes mellitus is projected to rise to 439 million globally, which represents 7.7% of the total adult population of the world’s adults several microRNAs are involved in -cell development and function, insulin secretion [71], and insulin resistance in the liver, skeletal muscle, and adipose tissue, which play an important role in glucose homeostasis and the pathogenesis of diabetes [72]. Altered microRNA expression also affects the progression of diabetic complications in the kidney, retina, and peripheral nerves. As each microRNA has the potential to regulate multiple genes in biological processes that include cell proliferation, differentiation, apoptosis, and development, it has been confirmed that the dysregulation of microRNAs affects many pathological pathways in diabetic complications [73, 74]. Decreased circulating miR- 126 was a significant predictor of DM. miR-15a, miR-29b, miR- 126, and miR-223 were decreased in the subjects with DM [75, 76]. In pancreatic -cell, islets, enriched miR-375 was increased in subjects with T2DM and modulated -cell function through several physiological mechanisms. miR-375 inhibits insulin secretion and transcription, maintains -cell mass, proliferation, and regeneration, and promotes embryonic pancreas development [77].
Role of miRNA in Cancer
Since the early stages of miRNA research, cancer has been the most prominent of human diseases with a clear role for miRNA regulation. The first evidence came from a study by Calin et al. in which they demonstrated a frequent deletion of miRNA genes miR15 and miR16 among 65% of B-cell chronic lymphocytic leukemia patients [78]. The deregulation of miR-125b, miR- 145, miR-21, and miR-155 expression was associated with the increased risk of breast cancer [79]. In addition, up-regulation of miR-155 and down-regulation of let-7a were correlated with poor survival of lung cancer patients [80]. Yu et al., reported low miRNA-34a levels in human bladder cancer tissues. Therefore, miRNA-34a is potential anti-angiogenic and anti-metastatic therapeutic target in bladder cancer patients [81]. Smits et al., showed that miRNA-101 inhibited proliferation, angiogenesis and migration of glioblastoma cells [82]. Moreover, Tang et al., showed that miRNA-101 was downregulated in nasopharyngeal carcinoma tissues and cell lines. They further showed that overexpression of miRNA-101 suppresses angiogenesis and lung metastasis [83]. miRNA-135a is a tumor suppressor, which is reported to be downregulated in human prostate and gallbladder cancers [84, 85]. Cheng et al. reported that miRNA-135a levels were downregulated in gastric cancer tissues and cell lines. They showed that miRNA-135a inhibited gastric cancer angiogenesis and metastasis [86].
miRNA Regulation on Immunity
MicroRNAs critically influence the development and responses of the immune system, but the contributing biological mechanisms are poorly characterized [87]. miRNA-155 is among the first miRNAs to be identified to play diverse roles in immunity and inflammation. It is generally believed that miRNA-155 is a multifunctional miRNA [88]. miRNA-155 has also been reported to regulate antigen presentation [89] and to regulate toll-like receptor (TLR) and cytokine signaling negatively. Rodriquez et al., showed that miR-155 is required for normal functioning of B and T lymphocytes, as well as dendritic cells [90]. Overexpression of the miR-17-92 cluster and miR-181 enhanced B-cell proliferation, while miR-150 regulated B-cell differentiation [91]. When overexpressed, miR-181 has been shown to decrease T-cell numbers [94], but enhance T-cell receptor signaling [90]. When T cells are activated, the miRNA expression profiles are altered [92, 93].
miRNA and Cerebrovascular Disease
miRNAs are crucial for the development of the nervous system. Recent studies have demonstrated that some miRNAs play important roles in the occurrence and development of ischemic cerebrovascular diseases such as stroke [94]. miR-126 could facilitate the angiogenesis following ischemia. Van Solingen et al., [95] firstly confirmed that miR-126 could facilitate the angiogenesis following ischemia. Bonauer et al., [96] also found that miR-92a could significantly inhibit the angiogenesis in vivo and in vitro. In addition, there was evidence that showed that miR- 21 may promote the proliferation of newly generated smooth muscle cells in the intima [97, 98] and downregulation of miR- 222 could facilitate the migration and proliferation of endothelial cells, which may increase the angiogenesis in the plaques [99, 100]. Cell apoptosis is an important pattern of cell death after stroke and plays important roles in the pathological processes following cerebral stroke. Some miRNAs (such as miR-497, -15a, and -21) have been found to be involved in the cell apoptosis after stroke. Besides apoptosis, miRNAs are also associated with brain edema after stroke [101]. Neurodegenerative diseases (ND) such as Parkinson’s disease (PD) and Alzheimer’s disease (AD) have placed substantial social-economic burdens on countries with aging populations. Notably, recent progresses from studies elucidating miRNA functions in NDs have shed newlight on disease pathogenesis and may lead to novel treatment strategies. For example, a systemic miRNA profiling in peripheral blood mononuclear cells from PD patients revealed miR-30b, miR-30c, and miR-26a to be associated with the susceptibility of the disease [102].
Conclusion
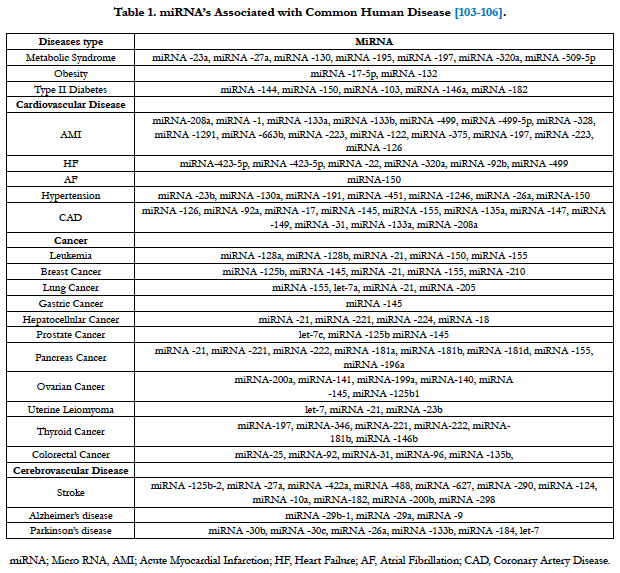
The effects of miRNAs on gene expression in different physiological processes and diseases have been determined. Recent studies shown that miRNA expression levels are important for many biological processes such as cell proliferation, differentiation, apoptosis, organogenesis and metabolism. So they serve as biomarkers and potential therapeutic targets and agents in various cancers and other diseases, e.g., cardiovascular disease, cerebrovascular disease and type II diabetes (table). Consequently it is thought that miRNAs will provided important benefits in the development of early diagnosis and treatment.
References
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116(2):281-97. PubMed PMID: 14744438.
- Feinberg MW, Moore KJ. MicroRNA regulation of atherosclerosis. Circ Res. 2016 Feb 19;118(4):703-20. doi: 10.1161/CIRCRESAHA.115.306300. PubMed PMID: 26892968.
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?. Nat Rev Genet. 2008 Feb;9(2):102-14. doi: 10.1038/nrg2290. PubMed PMID: 18197166.
- Kucher AN, Nazarenko MS, Markov AV, Koroleva IA, Barbarash OL. Variability of methylation profiles of CpG sites in microRNA genes in leukocytes and vascular tissues of patients with atherosclerosis. Biochemistry (Mosc). 2017 Jun;82(6):698-706. doi: 10.1134/S0006297917060062. PubMed PMID: 28601079.
- Kucher AN, Babushkina NP. Role of microRNA, genes involved in their biogenesis and functioning in the development of human disorders. Med. Genet. 2011;1(3):13.
- Vienberg S, Geiger J, Madsen S, Dalgaard LT. MicroRNAs in metabolism. Acta Physiol (Oxf ). 2017 Feb;219(2):346-361. doi: 10.1111/apha.12681. PubMed PMID: 27009502.
- Lee RC, Feinbaum RL, Ambros V. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993 Dec 3;75(5):843-54. PubMed PMID: 8252621.
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993 Dec 3;75(5):855-62. PubMed PMID: 8252622.
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000 Feb 24;403(6772):901-6. PubMed PMID: 10706289.
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin- 41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000 Apr;5(4):659-69. PubMed PMID: 10882102.
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000 Nov 2;408(6808):86-9. PubMed PMID: 11081512.
- Griffiths‐Jones S. The microRNA Registry. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D109-11. PubMed PMID: 14681370.
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008 Jan;36(Database issue):D154-8. Epub 2007 Nov 8. PubMed PMID: 17991681.
- Hastings ML, Krainer AR. Pre-mRNA splicing in the new millennium. Curr Opin Cell Biol. 2001 Jun;13(3):302-9. PubMed PMID: 11343900.
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003 Sep 25;425(6956):415-9. PubMed PMID: 14508493.
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004 Jan 2;303(5654):95-8. PubMed PMID: 14631048.
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003 Dec 15;17(24):3011-6. PubMed PMID: 14681208.
- Ardekani AM, Naeini MM. The Role of MicroRNAs in Human Diseases. Avicenna J Med Biotechnol. 2010 Oct;2(4):161-79. PubMed PMID: 23407304.
- Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008 Sep 1;79(4):581-8. doi: 10.1093/cvr/cvn156. PubMed PMID: 18550634.
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005 Mar;11(3):241-7. PubMed PMID: 15701730.
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004 Oct 13;23(20):4051-60. PubMed PMID: 15372072.
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, et al. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005 May 12;33(8):2697-706. PubMed PMID: 15891114.
- Herbert A. The four Rs of RNA-directed evolution. Nat Genet. 2004 Jan;36(1):19-25. PubMed PMID: 14702037.
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001 Feb 15;409(6822):860-921. PubMed PMID: 11237011.
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001 Feb 16;291(5507):1304-51. PubMed PMID: 11181995.
- Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: noncoding RNAs. B Biochim Biophys Acta. 2005 Sep 25;1756(1):65-75. PubMed PMID: 16125325.
- Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005 Apr 15;14 Spec No 1:R121-32. PubMed PMID: 15809264.
- Berezikov E, Plasterk RH. Camels and zebrafish, viruses and cancer: a microRNA update. Hum Mol Genet. 2005 Oct 15;14 Spec No. 2:R183-90. PubMed PMID: 16244316.
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005 Jul 15;122(1):6-7. PubMed PMID: 16009126.
- Klein ME, Impey S, Goodman RH. Role reversal: the regulation of neuronal gene expression by microRNAs. Curr Opin Neurobiol. 2005 Oct;15(5):507-13. PubMed PMID: 16150590.
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005 May 6;308(5723):833-8. PubMed PMID: 15774722.
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006 Mar;8(3):278-84. PubMed PMID: 16489342.
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola- Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005 Jun 16;435(7044):974-8. PubMed PMID: 15944714.
- Li M, Marin-Muller C, Bharadwaj U, Chow KH, Yao Q, Chen C. MicroRNAs: control and loss of control in human physiology and disease. World J Surg. 2009 Apr;33(4):667-84. doi: 10.1007/s00268-008-9836-x. PubMed PMID: 19030926.
- Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005 Dec 1;438(7068):685-9. PubMed PMID: 16258535.
- Rupaimoole R, Han HD, Lopez-Berestein G, Sood AK. MicroRNA therapeutics: principles, expectations, and challenges. Chin J Cancer. 2011 Jun;30(6):368-70. PubMed PMID: 21627858.
- Cheung WW, Mao P. Recent advances in obesity: genetics and beyond. ISRN Endocrinol. 2012;2012:536905. doi: 10.5402/2012/536905. PubMed PMID: 22474595.
- Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015 May;11(5):276-88. doi: 10.1038/ nrendo.2015.25. PubMed PMID: 25732520.
- Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A. 2008 Feb 26;105(8):2889-94. doi: 10.1073/pnas.0800178105. PubMed PMID: 18287052.
- Aranda JF, Madrigal-Matute J, Rotllan N, Fernández-Hernando C. Micro- RNA modulation of lipid metabolism and oxidative stress in cardiometabolic diseases. Free Radic Biol Med. 2013 Sep;64:31-9. doi: 10.1016/j. freeradbiomed.2013.07.014. PubMed PMID: 23871755.
- Hulsmans M, De Keyzer D, Holvoet P. MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis. FASEB J. 2011 Aug;25(8):2515-27. doi: 10.1096/fj.11-181149. PubMed PMID: 21507901.
- Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes. 2009 May;58(5):1050-7. doi: 10.2337/db08-1299. PubMed PMID: 19188425.
- Milagro FI, Miranda J, Portillo MP, Fernandez-Quintela A, Campión J, Martínez JA. High-throughput sequencing of microRNAs in peripheral blood mononuclear cells: identification of potential weight loss biomarkers. PLoS One. 2013;8(1):e54319. doi: 10.1371/journal.pone.0054319. PubMed PMID: 23335998.
- Carrer M, Liu N, Grueter CE, Williams AH, Frisard MI, Hulver MW, Bassel-Duby R, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci U S A. 2012 Sep 18;109(38):15330-5. PubMed PMID: 22949648.
- Prats-Puig A, Ortega FJ, Mercader JM, Moreno-Navarrete JM, Moreno M, Bonet N, et al. Changes in circulating microRNAs are associated with childhood obesity. J Clin Endocrinol Metab. 2013 Oct;98(10):E1655-60. doi: 10.1210/jc.2013-1496. PubMed PMID: 23928666.
- Li JH, Wang LM, Li YC, Bi YF, Jiang Y, Mi SQ, et al. Epidemiologic characteristics of dyslipidemia in Chinese adults 2010. Zhonghua Yu Fang Yi Xue Za Zhi. 2012 May;46(5):414-8. PubMed PMID: 22883727.
- Ross R. Atherosclerosis--an inflammatory disease. N Engl J Med. 1999 Jan 14;340(2):115-26. PubMed PMID: 9887164.
- Glueck CJ. Role of risk factor management in progression and regression of coronary and femoral artery atherosclerosis. Am J Cardiol. 1986 May 30;57(14):35G-41G. PubMed PMID: 3521250.
- Al Montasir A, Sadik MH. Acute myocardial infarction in a 28 year man with familial hypercholesterolemia. Indian J Med Sci. 2012 Mar-Apr;66(3-4):78-81. doi: 10.4103/0019-5359.110914. PubMed PMID: 23603625.
- Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003 Jun;111(12):1795- 803. PubMed PMID: 12813012.
- Ait-Oufella H, Taleb S, Mallat Z, Tedgui A. Recent advances on the role of cytokines in atherosclerosis. Arterioscler Thromb Vasc Biol. 2011 May;31(5):969-79. doi: 10.1161/ATVBAHA.110.207415. PubMed PMID: 21508343.
- Ramsey SA, Ramsey SA, Gold ES, Aderem A. A systems biology approach to understanding atherosclerosis. EMBO Mol Med. 2010 Mar;2(3):79-89. doi: 10.1002/emmm.201000063. PubMed PMID: 20201031.
- Zhang C. MicroRNAs in vascular biology and vascular disease. J Cardiovasc Transl Res. 2010 Jun;3(3):235-40. doi: 10.1007/s12265-010-9164-z. Pub- Med PMID: 20560045.
- Zhang C. MicroRNAs: role in cardiovascular biology and disease. Clin Sci (Lond). 2008 Jun;114(12):699-706. doi: 10.1042/CS20070211. PubMed PMID: 18474027.
- Cipollone F, Felicioni L, Sarzani R, Ucchino S, Spigonardo F, Mandolini C, et al. A unique microRNA signature associated with plaque instability in humans. Stroke. 2011 Sep;42(9):2556-63. doi: 10.1161/STROKEAHA. 110.597575. PubMed PMID: 21817153.
- Li T, Cao H, Zhuang J, Wan J, Guan M, Yu B, et al. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin Chim Acta. 2011 Jan 14;412(1-2):66-70. doi: 10.1016/j. cca.2010.09.029. PubMed PMID: 20888330.
- Raitoharju E, Lyytikäinen LP, Levula M, Oksala N, Mennander A, Tarkka M, et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 2011 Nov;219(1):211-7. doi: 10.1016/j.atherosclerosis.2011.07.020. PubMed PMID: 21820659.
- Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010 Sep 3;107(5):677-84. doi: 10.1161/CIRCRESAHA.109.215566. PubMed PMID: 20595655.
- Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013 Jan;123(1):11-8. doi: 10.1172/JCI62876. PubMed PMID: 23281405.
- Cong BH, Zhu XY, Ni X. The roles of microRNA-22 in myocardial infarction. Sheng Li Xue Bao. 2017 Oct 25;69(5):571-578. PubMed PMID: 29063105.
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a micro- RNA. Science. 2007 Apr 27;316(5824):575-9. PubMed PMID: 17379774.
- Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009 Nov;55(11):1944-9. doi: 10.1373/clinchem.2009.125310. PubMed PMID: 19696117.
- Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating micro- RNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010 Mar;31(6):659-66. doi: 10.1093/eurheartj/ehq013. PubMed PMID: 20159880.
- Gidlöf O, Andersson P, Van Der Pals J, Götberg M, Erlinge D. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology. 2011;118(4):217-26. doi: 10.1159/000328869. PubMed PMID: 21701171.
- Adachi T, Nakanishi M, Otsuka Y, Nishimura K, Hirokawa G, Goto Y, et al. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem. 2010 Jul;56(7):1183-5. doi: 10.1373/clinchem.2010.144121. PubMed PMID: 20395621.
- Cheng Y, Tan N, Yang J, Liu X, Cao X, He P, et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond). 2010 Apr 20;119(2):87-95. doi: 10.1042/CS20090645. PubMed PMID: 20218970.
- Cai WF, Liu GS, Lam CK, Florea S, Qian J, Zhao W, et al. Up‐regulation of micro‐RNA765 in human failing hearts is associated with post‐transcriptional regulation of protein phosphatase inhibitor‐1 and depressed contractility. Eur J Heart Fail. 2015 Aug;17(8):782-93. doi: 10.1002/ejhf.323. PubMed PMID: 26177627.
- Potus F, Ruffenach G, Dahou A, Thebault C, Breuils-Bonnet S, Tremblay È, Nadeau V, Paradis R, Graydon C, Wong R, Johnson I. Downregulation of miR-126 contributes to the failing right ventricle in pulmonary arterial hypertension. Circulation. 2015 Sep 8;132(10):932-43. doi: 10.1161/CIRCULATIONAHA. 115.016382. PubMed PMID: 26162916.
- Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010 Apr 2;106(6):1035-9. doi: 10.1161/CIRCRESAHA.110.218297. PubMed PMID: 20185794.
- Goren Y, Kushnir M, Zafrir B, Tabak S, Lewis BS, Amir O. Serum levels of microRNAs in patients with heart failure. Eur J Heart Fail. 2012 Feb;14(2):147-54. doi: 10.1093/eurjhf/hfr155. PubMed PMID: 22120965.
- Daneman D. Type 1 diabetes. Lancet. 2006 Mar 11;367(9513):847-58. PubMed PMID: 16530579.
- Chen L, Magliano DJ, Zimmet PZ. The worldwide epidemiology of type 2 diabetes mellitus - present and future perspectives. Nat Rev Endocrinol. 2011 Nov 8;8(4):228-36. doi: 10.1038/nrendo.2011.183. PubMed PMID: 22064493.
- Li X. MiR-375, a microRNA related to diabetes. Gene. 2014 Jan 1;533(1):1- 4. doi: 10.1016/j.gene.2013.09.105. PubMed PMID: 24120394.
- Shantikumar S, Caporali A, Emanueli C. Role of microRNAs in diabetes and its cardiovascular complications. Cardiovasc Res. 2012 Mar 15;93(4):583- 93. doi: 10.1093/cvr/cvr300. PubMed PMID: 22065734.
- Kantharidis P, Wang B, Carew RM, Lan HY. Diabetes complications: the microRNA perspective. Diabetes. 2011 Jul;60(7):1832-7. doi: 10.2337/ db11-0082. PubMed PMID: 21709278.
- Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010 Sep 17;107(6):810-7. doi: 10.1161/CIRCRESAHA.110.226357. PubMed PMID: 20651284.
- Mao Y, Mohan R, Zhang S, Tang X. MicroRNAs as pharmacological targets in diabetes. Pharmacol Res. 2013 Sep;75:37-47. doi: 10.1016/j. phrs.2013.06.005. PubMed PMID: 23810798.
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002 Nov 26;99(24):15524-9. PubMed PMID: 12434020.
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005 Aug 15;65(16):7065-70. PubMed PMID: 16103053.
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006 Mar;9(3):189-98. PubMed PMID: 16530703.
- Yu G, Yao W, Xiao W, Li H, Xu H, Lang B. MicroRNA-34a functions as an anti-metastatic microRNA and suppresses angiogenesis in bladder cancer by directly targeting CD44. J Exp Clin Cancer Res. 2014 Dec 31;33:779. doi: 10.1186/s13046-014-0115-4. PubMed PMID: 25551284.
- Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010 Dec;1(8):710-20. PubMed PMID: 21321380.
- Tang XR, Wen X, He QM, Li YQ, Ren XY, Yang XJ, et al. MicroRNA-101 inhibits invasion and angiogenesis through targeting ITGA3 and its systemic delivery inhibits lung metastasis in nasopharyngeal carcinoma. Cell Death Dis. 2017 Jan 19;8(1):e2566. doi: 10.1038/cddis.2016.486. PubMed PMID: 28102841.
- Kroiss A, Vincent S, Decaussin-Petrucci M, Meugnier E, Viallet J, Ruffion A, et al. Androgen-regulated microRNA-135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene. 2015 May 28;34(22):2846-55. doi: 10.1038/onc.2014.222. PubMed PMID: 25065599.
- Zhou H, Guo W, Zhao Y, Wang Y, Zha R, Ding J, et al. Micro RNA‐ 135a acts as a putative tumor suppressor by directly targeting very low density lipoprotein receptor in human gallbladder cancer. Cancer Sci. 2014 Aug;105(8):956-65. doi: 10.1111/cas.12463. PubMed PMID: 24903309.
- Cheng Z, Liu F, Zhang H, Li X, Li Y, Li J, et al. miR-135a inhibits tumor metastasis and angiogenesis by targeting FAK pathway. Oncotarget. 2017 May 9;8(19):31153-31168. doi: 10.18632/oncotarget.16098. PubMed PMID: 28415713.
- Gracias DT, Katsikis PD. MicroRNAs: key components of immune regulation. Adv Exp Med Biol. 2011;780:15-26. doi: 10.1007/978-1-4419-5632-3_2. PubMed PMID: 21842361.
- Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009 Jun;1792(6):497-505. doi: 10.1016/j.bbadis.2009.02.013. PubMed PMID: 19268705.
- Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006 Aug 15;103(33):12481-6. PubMed PMID: 16885212.
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007 Apr 27;316(5824):608-11. PubMed PMID: 17463290.
- Lu LF, Liston A. MicroRNA in the immune system, microRNA as an immune system. Immunology. 2009 Jul;127(3):291-8. doi: 10.1111/j.1365-2567.2009.03092.x. PubMed PMID: 19538248.
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004 Jan 2;303(5654):83-6. PubMed PMID: 14657504.
- Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007 Apr 6;129(1):147-61. PubMed PMID: 17382377.
- Yan H, Fang M, Liu XY. Role of microRNAs in stroke and poststroke depression. Sci World J. 2013 Dec 2;2013:459692. doi: 10.1155/2013/459692. PubMed PMID: 24363618.
- van Solingen C, Seghers L, Bijkerk R, Duijs JM, Roeten MK, van Oeveren‐ Rietdijk AM, et al. Antagomir‐mediated silencing of endothelial cell specific microRNA‐126 impairs ischemia‐induced angiogenesis. J Cell Mol Med. 2009 Aug;13(8A):1577-85. doi: 10.1111/j.1582-4934.2008.00613.x. PubMed PMID: 19120690.
- Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009 Jun 26;324(5935):1710-3. doi: 10.1126/science.1174381. PubMed PMID: 19460962.
- Weber M, Baker MB, Moore JP, Searles CD. MiR-21 is induced in endothelial cells by shear stress and modulates apoptosis and eNOS activity. Biochem Biophys Res Commun. 2010 Mar 19;393(4):643-8. doi: 10.1016/j. bbrc.2010.02.045. PubMed PMID: 20153722.
- Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007 Jun 8;100(11):1579-88. PubMed PMID: 17478730.
- Dentelli P, Rosso A, Orso F, Olgasi C, Taverna D, Brizzi MF. micro- RNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler Thromb Vasc Biol. 2010 Aug;30(8):1562-8. doi: 10.1161/ATVBAHA.110.206201. PubMed PMID: 20489169.
- Wu YH, Hu TF, Chen YC, Tsai YN, Tsai YH, Cheng CC, et al. The manipulation of microRNA-gene regulatory networks by KSHV induces endothelial cell motility. Blood. 2011 Sep 8;118(10):2896-905. doi: 10.1182/blood-2011-01-330589. PubMed PMID: 21715310.
- Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009 Apr;29(4):675-87. doi: 10.1038/jcbfm.2008.157. PubMed PMID: 19142192.
- Martins M, Rosa A, Guedes LC, Fonseca BV, Gotovac K, Violante S, et al. Convergence of miRNA expression profiling, α-synuclein interacton and GWAS in Parkinson's disease. PLoS One. 2011;6(10):e25443. doi: 10.1371/journal.pone.0025443. PubMed PMID: 22003392.
- Iacomino G, Siani A. Role of microRNAs in obesity and obesity-related diseases. Genes Nutr. 2017 Sep 25;12:23. doi: 10.1186/s12263-017-0577-z. PubMed PMID: 28974990.
- Chien HY, Lee TP, Chen CY, Chiu YH, Lin YC, Lee LS, et al. Circulating microRNA as a diagnostic marker in populations with type 2 diabetes mellitus and diabetic complications. J Chin Med Assoc. 2015 Apr;78(4):204-11. doi: 10.1016/j.jcma.2014.11.002. PubMed PMID: 25529478.
- Li M, Zhang J. Circulating microRNAs: potential and emerging biomarkers for diagnosis of cardiovascular and cerebrovascular diseases. Biomed Res Int. 2015;2015:730535. doi: 10.1155/2015/730535. PubMed PMID: 26180810.
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199-227. doi: 10.1146/annurev.pathol.4.110807.092222. PubMed PMID: 18817506.