Differential Expression Profiles and Functional Analysis of Circular RNAs of Sinonasal Inverted Papillomas
Qiang Zhang1#, Xiaoyue Zhen2#, Zhiyong Yue1, Zhaoyang Cui1, Ling Guo1, Deheng Luan1, Kun Gao1*, Xuanchen Zhou1*
1 Department of Otorhinolaryngology Head and Neck Surgery, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China.
2 Minimally Invasive Urology Center, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China.
# These authors contributed equally to this paper.
*Corresponding Author
Kun Gao, Xuanchen Zhou,
Department of Otorhinolaryngology Head and Neck Surgery, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, China.
Tel: +8618769786627
E-mail:gaokunsy@163.com/xuanchenzhou@163.com
Received: November 07, 2022; Accepted: June 27, 2023; Published: July 11, 2023
Citation: Qiang Zhang, Xiaoyue Zhen, Zhiyong Yue, Zhaoyang Cui, Ling Guo, Deheng Luan, Kun Gao, Xuanchen Zhou. Differential Expression Profiles and Functional Analysis
of Circular RNAs of Sinonasal Inverted Papillomas. Int J Clin Exp Otolaryngol. 2023;7(1):143-150.
Copyright:Kun Gao, Xuanchen Zhou©2023. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Aim: To find out the differential expression profiles of circular RNAs (circRNAs) between sinonasal inverted papilloma (SNIP) and normal nasal mucosa tissue, and then explore their possible functions.
Methods: The circRNA microarray experiment was carried out to find the differential expression profiles in SNIP and normal nasal mucosa tissues in six patients. The functional analysis was used to better understand the biological functions of dysregulated circRNAs. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to validation of the microarray data. The functional analysis was used to better understand the biological functions of dysregulated circRNAs. Co-expression network and circRNA-miRNA-mRNA network analyses were also performed to study molecular interactions.
Results: 4264 circRNAs were significantly dysregulated (fold change≥2 , P<0.05) which contain 1567 up-regulated and 2697 down-regulated. Functional analysis found that these dysregulated circRNAs mainly contribute to regulation of cell growth, proliferation differentiation and degradation. The qRT-PCR confirmed has_circ_0026421 up-regulation and has_ circ_0027511 down-regulation in SNIP tissue. The top two MREs of hsa_circ_0027511 are hsa_miR-548v and hsa_miR-557, and the top two miRNA response elements (MREs) of hsa_circ_0026421 are hsa_miR-6512-3p and hsa_miR-6722-3p.
Conclusion: CircRNAs may play an important role in SNIP. Has_circ_0026421 and has_circ_0027511 could be used as promising markers. These results of circRNAs can provide important theoretical foundations for further studies on the function of circRNAs in SNIP, and provide new ideas for the diagnosis and treatment of SNIP.
2.Introduction
3.Materials and Methods
4.Results
5.Discussion
6.Conclusion
7.References
Keywords
Sinonasal Inverted Papilloma; Circular RNAs, Gene Expression Profile; Microarray; ceRNA.
Introduction
Sinonasal inverted papilloma (SNIP) is one of the most common
benign tumors of sinonasal cavities and it accounts for 0.5-4%
of sinonasal tumors and about 70% sinonasal papilloma [1]. It
is originated from ectodemal schneiderian membrane of nasal
cavity or paranasal sinuses [2, 3]. Compared with nasal polyps,
the lesions of SNIP are firmer, bulkier and lack translucency. Its
main clinicopathological features include the tendency to recur,
easily malignant transformation and its aggressive growth which
can cause local invasion and bone destruction [4, 5]. Nasal obstruction,
rhinorrhoea, epistaxis, headache and anosmia are main
symptoms of SNIP. The optimal treatment for SNIP is surgery
which aimed at removal the tumors completely. The etiology and
pathogenesis of SNIP are still unclear. Study about molecular
mechanisms can help us better understand the development of
SNIP and may provide a better treatment for SNIP.
Noncoding RNAs (ncRNAs) are RNAs that do not encode proteins,
which including small nuclear RNAs, transfer RNAs and
ribosomal RNAs, as well as the more recently discovered long
noncoding RNAs (lncRNAs), circular RNAs (circRNAs) and
microRNAs (miRNAs), and they have critical regulatory roles in
many biological functions [6, 7]. CircRNAs are a class of ncR-NAs which were first discovered in human in 1986 [8]. They
are present in the cytoplasm of eukaryotic cells and are singlestranded
circular molecules formed by pre-mRNA back-splicing
through variable shearing processes [9-12]. CircRNAs are more
stable compared with linear RNAs because they are covalently
closed loop structures with no 5’ to 3’ polarity and polyadenylation
which make them resistance to RNA exonucleases [13, 14].
Potential biological functions of circRNAs is still unclear, but increased
researches reported that circRNAs can act as competing
endogenous RNAs (ceRNAs) of miRNAs via miRNA response
elements (MREs)[15] which can regulate transcription and gene
expression [16]. Many studies have found that circRNA play an
important role in many life processes including aging, tissue development,
insulin secretion, and cell apoptosis [17]. Also, circRNAs
are related to several diseases, such as cancer, autoimmune
disease, cardiovascular diseases and neurological disorders [18,
19]. However, few research focus on the role of circRNAs in
SNIP and the expression profiles of circRNAs in SNIP are still
undiscovered. In this study, we explored the differential expression
profiles between the SNIP and normal nasal mucosa tissue
using RNA microarray. Then, quantitative real- time reverse transcription
PCR (qRT-PCR) was used to validate the selected differentially
expressed circRNAs.
Materials and Methods
Patients and Sample Collection
SNIP specimens and normal nasal mucosa tissues outside the tumor
were obtained from eight patients undergoing endoscopic
resection of inverted papilloma at Department of Otorhinolaryngology
of Shandong Provincial Hospital Affiliated to Shandong
First Medical University. The specimens were collected between
June 2020 and May 2021. All SNIP cases were confirmed by postoperative
pathology. The exclusion criteria were as follows: recurrent
SNIP, combined with other neoplastic lesions of nasal cavity
and paranasal sinus, accompanied by systemic malignant tumors
or immune diseases. The basic clinical characteristics of eight
patients with SNIP are shown in table 1. The tumor tissue and
normal tissue of each patient were used as controls. Besides, six
pairs of samples (SNIP_1-6) were used for microarray analysis,
and three pairs of samples (SNIP_6-8, including one pair used for
microarray) were used by qRT-PCR for microarray data verification.
The research was improved by the institutional Ethics Committee
of Shandong Provincial Hospital Affiliated to Shandong
First Medical University.
RNA extraction and microarray analysis
Total RNA was isolated using RNeasy Total RNA Isolation Kit
(Qiagen, GmBH, Germany)/ TRIzol reagent (Life technologies,
Carlsbad, CA, US) according to the manufacturer’s instructions,
and purified by using a RNeasy Mini Kit (Qiagen, GmBH, Germany).
Total RNA was checked for a RIN number to inspect RNA
integration by an Agilent Bioanalyzer 2100 (Agilent technologies,
Santa Clara, CA, US). RNA samples of each group were then
used to generate biotinylated cRNA targets for the Sino Human
ceRNA array V3.0. The biotinylated cRNA targets were then hybridized
with the slides. After hybridization, slides were scanned
on the Agilent Microarray Scanner (Agilent technologies, Santa
Clara, CA, US). Data were extracted with Feature Extraction software
10.7 (Agilent technologies, Santa Clara, CA, US). Raw data
were normalized by Quantile algorithm, R package “limma”. The
microarray experiments were performed by following the protocol
of Agilent technologies Inc at Sinotech Genomics Corporation.
Genes with a fold change of at least 2 were selected for
further analysis. Heatmap plots were done by a R package “pheatmap”
of the target genes.
Functional analysis by GO and KEGG pathway
Gene ontology (GO) (www.geneontology.org) is a kind of ontology
which is widely used in the bioinformatics field. GO categories
were divided into three parts according to its function: biological
process (BP) and cellular component (CC), molecular function
(MF). It was used to explore the functions of up-regulated and
down-regulated genes. Kyoto encyclopedia of genes and genomes
(KEGG) (www.kegg.jp) pathway analysis is a frequently used
analysis of gene function and genome information database and
it can help researchers to find out gene and expression information.
KEGG enrichment analysis of dysregulated genes can find
out differentially enriched pathways, which is helpful to identify
the functions of these dysregulated genes in the biological regulation
pathway. GO/pathway enrichment analysis were done use
Fisher's exact test by a R package “clusterProfiler” of the target
genes. GO categories/Pathway with Fisher's exact test P values <
0.05 were selected.
Quantitative real time-PCR
Quantitative real time-PCR (qRT-PCR) was carried out by using
LightCycler 480 (Roche, Shanghai, China) system to quantitate the levels of three up-regulated circRNAs (has_circ_0025388,
has_circ_0014213, has_circ_26421) and three down-regulated
circRNAs(has_circ_0027511, has_circ_0081375, has_
circ_0013507). The qRT-PCR was conducted by the instructions
of the SYBR® Green Premix Pro Taq HS qPCR Kit (AG, China).
The primers sequences of circRNAs for qRT-PCR validation are
shown in Table 2. The relative expression levels of circRNAs
were presented using 2-ΔΔCt method.
Co-expression network
Co-expression network is to calculate the co-expression relationship between genes according to the dynamic change of gene expression signal value, obtain the expression regulation relationship and regulation direction between genes, and then construct the gene expression regulation network. In order to better understood the function of circRNAs and the relationship between circRNA and mRNA, we calculated the co-expression relationship between circRNA and mRNA, so as to construct the gene expression regulation network. By using the co-expression network, we can analyze the gene regulation ability and obtain the core regulatory genes that the samples change with the experiment. We construct the co-expression network between circRNAs and mRNAs using Cytoscape.
Competing endogenous RNA (ceRNA) network construction
Recent studies have shown that circRNA molecules are rich in microRNA binding sites, which can act as miRNA sponge in cells, and further relieve the inhibition of miRNA on its target genes, and increase the expression level of the target genes. This mechanism is called the competitive endogenous RNA (ceRNA) mechanism. The ceRNA network base on this hypothesis which reveals a new mechanism of RNA interaction that ceRNA (mRNA, lncRNA, circRNA) can regulate each other's expression by competitive binding miRNA [20, 21]. CeRNA analysis is based on gene expression values. Through regression model analysis and seed sequence matching, a regulatory network for sponge adsorption of microRNA is established to find the core ceRNA. Interactions of circRNAs with potential miRNA response elements (MREs) were assessed with Cytoscape software. Then we constructed the circRNA-miRNA-mRNA network of the two circRNAs (has_ circ_0026421 and has_circ_0027511) which verified in qRT-PCR.
Statistical analysis
The data were expressed as mean ± standard deviation (SD). The differences between the two groups of data were compared by t-test using SPSS 24.0 software. p<0.05 indicated statistically significance.
Results
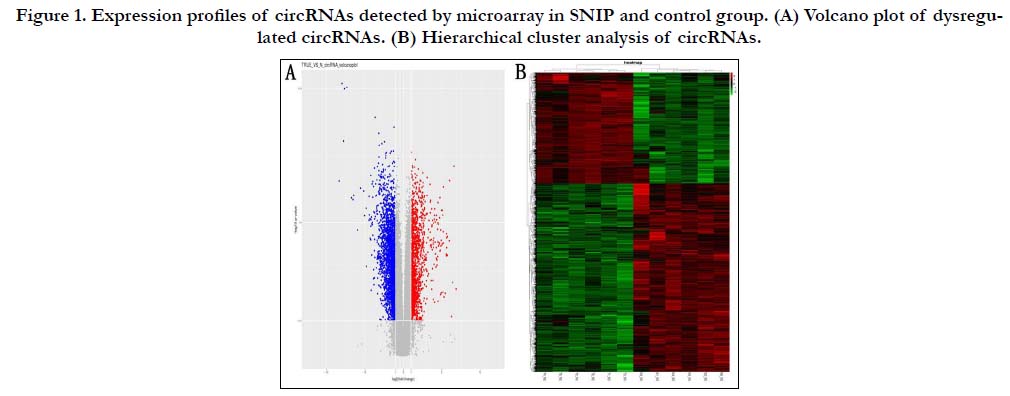
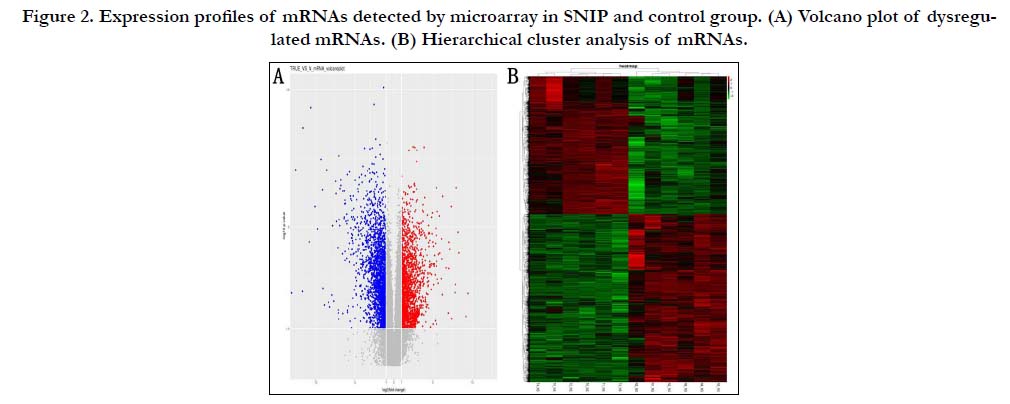
Dysregulatated circRNA and mRNA
In order to explore the dysregulated circRNA and mRNA in
SNIP, the microarray analysis was performed using SNIP samples
and normal nasal mucosa samples. According to the results of
circRNA microarray analysis, 4264 circRNAs were significantly
dysregulated (fold change≥2, p<0.05) which contain 1567 upregulated
and 2697 down-regulated (Fig. 1). Based on the result
of mRNA microarray analysis, 4194 mRNAs were significantly
dysregulated (fold change≥2, p<0.05) , including 1894 up-regulated
and 2300 down-regulated (Fig. 2). Of which the top 10 upregulated
and down-regulated circRNAs were shown in Table 3.
Figure 1. Expression profiles of circRNAs detected by microarray in SNIP and control group. (A) Volcano plot of dysregulated circRNAs. (B) Hierarchical cluster analysis of circRNAs.
Figure 2. Expression profiles of mRNAs detected by microarray in SNIP and control group. (A) Volcano plot of dysregulated mRNAs. (B) Hierarchical cluster analysis of mRNAs.
Table 3. Top 10 up-regulated and down-regulated (fold-change≥2 & p<0.05) circRNAs based on fold change in SNIP samples.
Function analysis of dysregulated circRNAs
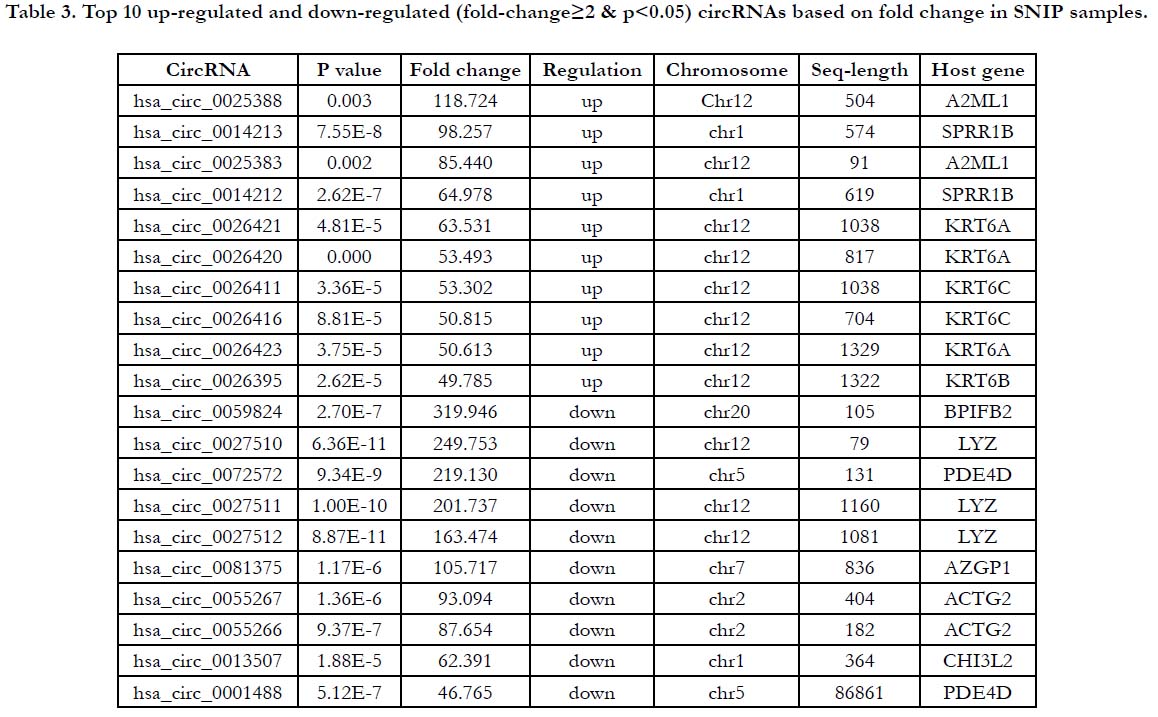
GO analysis were carried in order to analyze which gene functions are related to differentially expressed circRNAs. The top 3 enriched GO terms of dysregulated circRNAs were “cellular process” “single-organism process” “biological regulation” in BP; “cell” “cell part” “organelle” in CC; “binding” “catalytic activity” “molecular function regulator” in MF (Fig. 3A). Similarly, KEGG pathway analysis was used to explore which pathways were related to dysregulated circRNAs. And the results of top 30 pathways between SNIP samples and normal nasal mucosa samples were shown in Fig.3B. The focal adhesion pathway, apelin signaling pathway and ECM-receptor interaction pathway were enriched in dysregulated circRNAs. These results suggested that these pathways related to regulation of cell growth, proliferation differentiation and degradation may contribute to the pathogenesis of SNIP.
Validation of circRNA expression
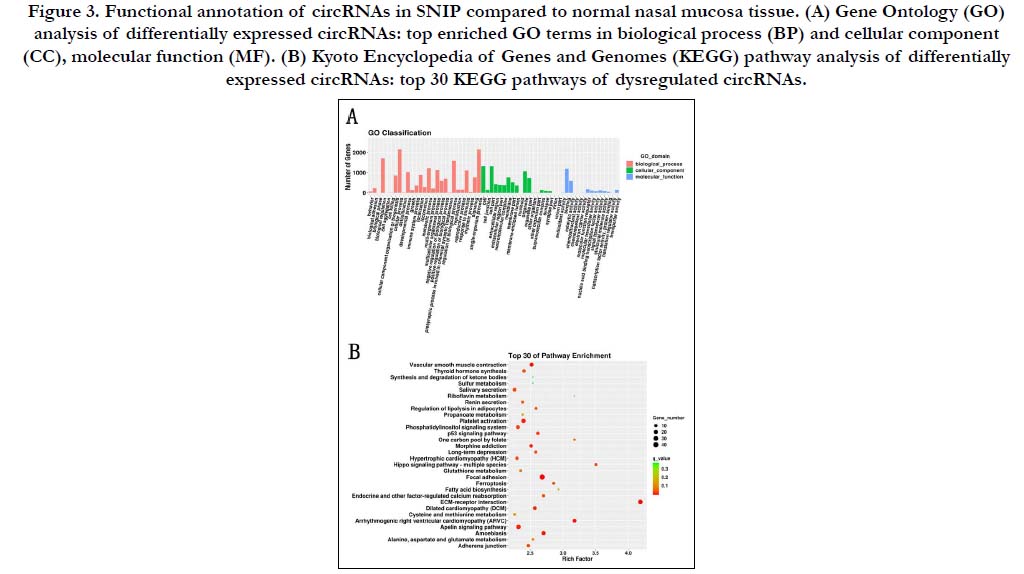
Six circRNAs including three up-regulated and three downregulated which were chosen from microarray data were validated using the qRT-PCR. Of which has_circ_0014213 and has_ circ_0025388 failed in qRT-PCR analysis because of nonspecific products. The expression trend of has_circ_0013507 was inconsistent. Among the remaining three circRNAs, there was no significant difference in the relative expression of has_circ_0081375 between normal and SNIP tissues. While the results of other two circRNAs (has_circ_0026421 and has_circ_0027511) were consistent with the microarray, and the relative expression differences were statistically significant (Fig. 4).
Co-expression network construction of circRNAs and mRNAs
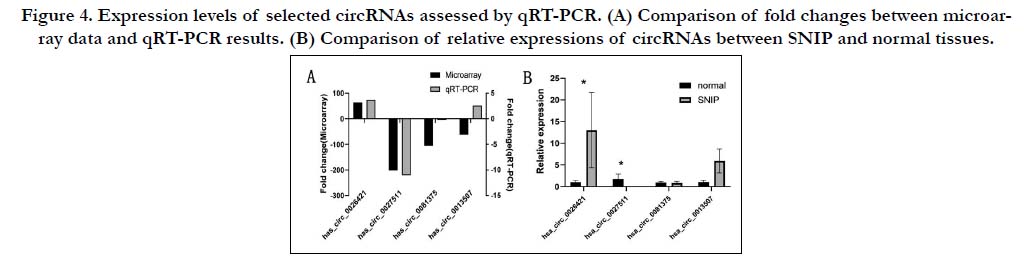
According to the microarray data and calculation results of a Pearson’s correlation coefficient, numerous coordinately expressed genes’ networks were construction in SNIP tissue compared with the normal nasal mucosa. By analyzing the functional analysis (GO analysis and KEGG pathway analysis) of the top 20 circRNAs (ten most up-regulated and ten down-regulated), we selected two pathways of Rap 1 signaling pathway and cAMP signaling pathway. Based on these two pathways, we constructed a circRNA-mRNA co-expression network of the ten most up-regulated and down-regulated circRNAs (Fig. 5). The results shows that circRNAs work together with other circRNAs and mRNAs rather than act alone.
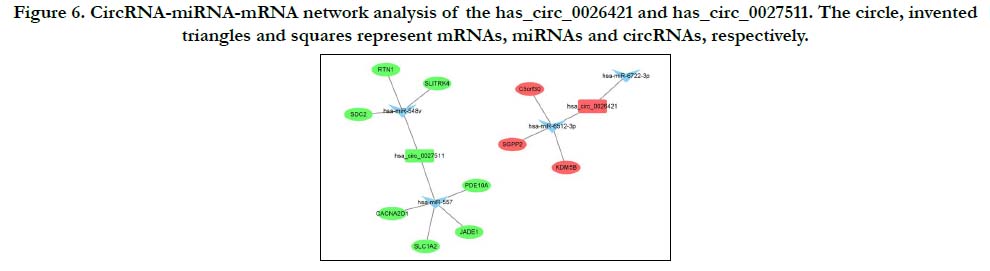
ceRNA network construction of circRNA-miRNA-mRNA
CircRNA molecules are rich in miRNA binding sites and play the role of miRNA sponge in cells, so as to relieve the inhibition of miRNA on its target genes and increase the expression level of target genes. This mechanism is called ceRNA mechanism. Considering microarray analysis and verification results, has_ circ_0026421 and has_circ_0027511 were expressed significantly differentially in SNIP. In order to explore the possible pathways of the verified circRNAs, based on the two pathways mentioned above (Rap 1 signaling pathway and cAMP signaling pathway), we constructed circRNA-miRNA-mRNA network of these two selected circRNAs which was shown in Fig. 6. The results showed that the top two MREs of hsa_circ_0027511 are hsa_miR-548v and hsa_miR-557, and the top two MREs of hsa_circ_0026421 are hsa_miR-6512-3p and hsa_miR-6722-3p.
Figure 3. Functional annotation of circRNAs in SNIP compared to normal nasal mucosa tissue. (A) Gene Ontology (GO) analysis of differentially expressed circRNAs: top enriched GO terms in biological process (BP) and cellular component (CC), molecular function (MF). (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed circRNAs: top 30 KEGG pathways of dysregulated circRNAs.
Figure 4. Expression levels of selected circRNAs assessed by qRT-PCR. (A) Comparison of fold changes between microarray data and qRT-PCR results. (B) Comparison of relative expressions of circRNAs between SNIP and normal tissues.
Figure 5. CircRNA-mRNA co-expression network analysis of the ten most up-regulated and down-regulated circRNAs. The circles and squares represent mRNAs and circRNAs, respectively.
Figure 6. CircRNA-miRNA-mRNA network analysis of the has_circ_0026421 and has_circ_0027511. The circle, invented triangles and squares represent mRNAs, miRNAs and circRNAs, respectively.
Discussion
Sinonasal inverted papilloma is a benign epithelial tumor, which
originates from Schneider mucosa of nasal cavity and paranasal sinus
[22]. Although the exact etiology of SNIP remains unknown,
it is a multifactorial disease that may be related to human papilloma
virus (HPV) infection, chronic inflammation, allergy and
occupational exposure [23]. At present, the diagnosis, treatment
and follow-up of SNIP are mainly based on imaging, pathology
and endoscopic technology. But there was still a high recurrence
rate and malignant transformation rate after operation. With the
development of molecular biology technology, the research on
the mechanism of SNIP occurrence, development, proliferation
and malignant transformation has made more in-depth progress
at home and abroad. Li et al.[24] found that the overexpression
of ErbB1 and ErbB2 in SNIP epithelial cells may play an important
role in the pathogenesis and progress of SNIP. ErbB1 and
erbB2 may be new targets for the treatment of SNIP. Yasukawa et
al.[25] confirmed that kirsten rat sarcoma viral oncogene (KRAS)
gene is a key factor in predicting the malignant transformation of
SNIP in Japanese patients. Lin et al.[26] found that Stathmin was
up-regulated in SNIP patients by immunohistochemical staining.
Yamashita et al.[27] proposed that there was a significant correlation
between serum squamous cell carcinoma antigen (SCCA)
level and SNIP size, and the increase of SCCA was a significant
predictor of tumor size. These studies may be helpful to explore
the key genes affecting SNIP and provide new ideas and favorable
evidence for the diagnosis and treatment of SNIP. However, most
studies have not been used in clinic because of insufficient sample
size, geographical limitations and inability to identify key factors.
As is known to all, many human genome sequences do not encode
any protein, and studies found that non-coding RNA accounts
for almost 95% of the total RNA sequences transcribed
from the human genome, including circular RNA[28]. At the time
when the circRNA was first be discovered, it was considered to
be a low-abundance RNA molecule which formed by mis-splicing
of exons when transcription [29]. With the development of technology
of bioinformatics and RNA sequencing, circRNAs are be
found present widely in the eukaryotic cells and play an key role
in regulating gene expression at the post-transcriptional level. So
far, they have been reported to be related to regulating immunity,
inflammations, and cell proliferation and they are thought
to play essential roles in a number of diseases [30-33]. Based on
the above research results, we considered that circRNAs may play
an important role in SNIP. However, accurate expression profile
of circRNAs and their possible functions in SNIP have not been
reported so far. In this present study, we try to explore the expression
profile of circRNA in SNIP and analyze its potential
functions.
Firstly, we explored the expression profiles of circRNAs of SNIP
tissue and normal nasal mucosa extracted from six patient with
SNIP when undergoing endoscopic sinus surgery. As mentioned
earlier, a total of 4264 circRNAs were significantly dysregulated
(fold change≥2 , p<0.05) which contain 1567 up-regulated and
2697 down-regulated. Then, in order to confirm the accuracy of
the microarray results, we selected 6 abnormally expressed circRNAs
(including 3 up-regulated and 3 down-regulated), which were
verified by qRT-PCR in three groups of tumor and adjacent normal
tissue specimens. Three of them were confirmed to show the
same trend of circRNAs in the qRT-PCR and microarray, in which,
relative expression differences of two circRNAs were statistically
significant, including one was up-regulated(has_circ_0026421)
and the other was down-regulated (has_circ_0027511).
Then, we constructed the co-expression network of circRNAmRNA
and circRNA-miRNA-mRNA network to obtain the relationship
between circRNAs and mRNAs. And we focused on
the analysis of ten most up-regulated and down-regulated circRNAs,
especially the two verified circRNAs (has_circ_0026421 and
has_circ_0027511). Results showed that the top two MREs of
hsa_circ_0027511 are hsa_miR-548v and hsa_miR-557, and the
top two MREs of hsa_circ_0026421 are hsa_miR-6512-3p and hsa_miR-6722-3p. These finding can enable us to further understand
the pathogenesis of SNIP.
The GO and KEGG analyses were performed to understand the
biological functions of dysregulatated circRNA. The three most
enriched GO terms were “cellular process” “single-organism process”
and “biological regulation”. And KEGG pathways were
enriched “focal adhesion pathway”, “apelin signaling pathway”
and “ECM-receptor interaction pathway”. These pathways were
related to regulation of cell growth, proliferation differentiation
and degradation [34-36]. Thus, these current findings mean the
dysregulated circRNAs might contribute to SNIP via these pathways
and through these biological processes. Further researches
will focus on the action sites and regulation modes of these pathways
and biological processes.
In the future study, we will focus more on has_circ_0026421 and
has_circ_0027511, and more experiments will be used to further
evaluate the role potential role of has_circ_0026421 and has_
circ_0027511 functioned as ceRNA in SNIP.
Conclusion
In conclusion, this study demonstrated that circRNAs are aberrantly
expressed in SNIP. Has_circ_0026421 was significantly
up-regulated and has_cirC_0027511 were significantly down-regulated
in SNIP tissues, which suggested that they may be novel
biomarker for SNIP diagnosis and targeted therapy. In the future,
more studies are needed to investigate the potential of circRNAs
as biomarkers for SNIP diagnosis and targeted therapy.
Funding
This work was supported by the Shandong Province Medical
and Health Technology Development Plan Project (grant
no.2019WS500), the Key Technology Research and Development
Program of Shandong (Grant no. 2019GSF108257 and
2018GSF118192).
References
- Wood JW, Casiano RR. Inverted papillomas and benign nonneoplastic lesions of the nasal cavity. Am J Rhinol Allergy. 2012 Mar-Apr;26(2):157-63. PubMed PMID: 22487294.
- Melroy CT, Senior BA. Benign sinonasal neoplasms: a focus on inverting papilloma. Otolaryngol Clin North Am. 2006 Jun;39(3):601-17. PubMed PMID: 16757234.
- Vrabec DP. The inverted Schneiderian papilloma: a 25-year study. Laryngoscope. 1994 May;104(5 Pt 1):582-605. PubMed PMID: 8189990.
- Lisan Q, Laccourreye O, Bonfils P. Sinonasal inverted papilloma: From diagnosis to treatment. Eur Ann Otorhinolaryngol Head Neck Dis. 2016 Nov;133(5):337-341. PubMed PMID: 27053431.
- Karligkiotis A, Lepera D, Volpi L, Turri-Zanoni M, Battaglia P, Lomardi D, et al. Survival outcomes after endoscopic resection for sinonasal squamous cell carcinoma arising on inverted papilloma. Head Neck. 2016 Nov;38(11):1604-1614. PubMed PMID: 27152722.
- Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta. 2016 Jan;1859(1):128-38. PubMed PMID: 26073320.
- Salzman J. Circular RNA Expression: Its Potential Regulation and Function. Trends Genet. 2016 May;32(5):309-316. PubMed PMID: 27050930.
- Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986 Oct 9-15;323(6088):558-60. PubMed PMID: 2429192.
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013 Sep 26;51(6):792-806. Pub- Med PMID: 24035497.
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife. 2015 Jun 9;4:e07540. PubMed PMID: 26057830.
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013 Mar 21;495(7441):333-8. PubMed PMID: 23446348.
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9(9):e1003777. PubMed PMID: 24039610.
- Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, et al. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015 Sep 1;365(2):141-8. PubMed PMID: 26052092.
- Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006 May 8;34(8):e63. PubMed PMID: 16682442.
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013 Mar 21;495(7441):384-8. PubMed PMID: 23446346.
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016 Sep 1;37(33):2602-11. PubMed PMID: 26802132.
- Li CY, Ma L, Yu B. Circular RNA hsa_circ_0003575 regulates oxLDL induced vascular endothelial cells proliferation and angiogenesis. Biomed Pharmacother. 2017 Nov;95:1514-1519. PubMed PMID: 28946214.
- Verduci L, Tarcitano E, Strano S, Yarden Y, Blandino G. CircRNAs: role in human diseases and potential use as biomarkers. Cell Death Dis. 2021 May 11;12(5):468. PubMed PMID: 33976116.
- Wang M, Yu F, Wu W, Zhang Y, Chang W, Ponnusamy M, et al. Circular RNAs: A novel type of non-coding RNA and their potential implications in antiviral immunity. Int J Biol Sci. 2017 Nov 2;13(12):1497-1506. PubMed PMID: 29230098.
- Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther. 2018 Jul;187:31-44. PubMed PMID: 29406246.
- Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018 Apr 7;17(1):79. PubMed PMID: 29626935.
- Lund VJ, Stammberger H, Nicolai P, Castelnuovo P, Beal T, Beham A, et al. European position paper on endoscopic management of tumours of the nose, paranasal sinuses and skull base. Rhinol Suppl. 2010 Jun 1;22:1-143. PubMed PMID: 20502772.
- Gupta R, Rady PL, Sikora AG, Tyring SK. The role of human papillomavirus in the pathogenesis of sinonasal inverted papilloma: a narrative review. Rev Med Virol. 2021 May;31(3):e2178. PubMed PMID: 33048407.
- Sahnane N, Ottini G, Turri-Zanoni M, Furlan D, Battaglia P, Karligkiotis A, et al. Comprehensive analysis of HPV infection, EGFR exon 20 mutations and LINE1 hypomethylation as risk factors for malignant transformation of sinonasal-inverted papilloma to squamous cell carcinoma. Int J Cancer. 2019 Mar 15;144(6):1313-1320. PubMed PMID: 30411788
- Yasukawa S, Kano S, Hatakeyama H, Nakamaru Y, Takagi D, Mizumachi T, et al. Genetic mutation analysis of the malignant transformation of sinonasal inverted papilloma by targeted amplicon sequencing. Int J Clin Oncol. 2018 Oct;23(5):835-843. PubMed PMID: 29779136.
- Lin H, Lin D, Xiong XS. Roles of human papillomavirus infection and stathmin in the pathogenesis of sinonasal inverted papilloma. Head Neck. 2016 Feb;38(2):220-4. PubMed PMID: 25224680.
- Yamashita Y, Uehara T, Hasegawa M, Deng Z, Matayoshi S, Kiyuna A, et al. Squamous cell carcinoma antigen as a diagnostic marker of nasal inverted papilloma. Am J Rhinol Allergy. 2016 Mar-Apr;30(2):122-7. PubMed PMID: 26877539.
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013 Feb;19(2):141-57. PubMed PMID: 23249747.
- Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. 1993 Jan;7(1):155-60. PubMed PMID: 7678559.
- Zhang L, Han B, Wang J, Liu Q, Kong Y, Jiang D, et al. Differential expression profiles and functional analysis of circular RNAs in children with fulminant myocarditis. Epigenomics. 2019 Aug;11(10):1129-1141. PubMed PMID: 31198064.
- Qiao M, Ding J, Yan J, Li R, Jiao J, Sun Q. Circular RNA Expression Profile and Analysis of Their Potential Function in Psoriasis. Cell Physiol Biochem. 2018;50(1):15-27. PubMed PMID: 30278433.
- Guo J, Han B, Wang J, Zhang L, Chen N, Sun W, et al. The differential expression and potential roles of circular RNAs in children with anti-NMDA receptor encephalitis. J Neuroimmunol. 2020 Nov 15;348:577381. PubMed PMID: 32911360.
- Zhuo CJ, Hou WH, Jiang DG, Tian HJ, Wang LN, Jia F, et al. Circular RNAs in early brain development and their influence and clinical significance in neuropsychiatric disorders. Neural Regen Res. 2020 May;15(5):817- 823. PubMed PMID: 31719241.
- Alanko J, Ivaska J. Endosomes: Emerging Platforms for Integrin-Mediated FAK Signalling. Trends Cell Biol. 2016 Jun;26(6):391-398. PubMed PMID: 26944773.
- Antushevich H, Wójcik M. Review: Apelin in disease. Clin Chim Acta. 2018 Aug;483:241-248. PubMed PMID: 29750964.
- Bao Y, Wang L, Shi L, Yun F, Liu X, Chen Y, et al. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell Mol Biol Lett. 2019 Jun 6;24:38. Pub- Med PMID: 31182966.