Knowledge, Attitude And Perception On Genomic Markers In Oral Potentially Malignant Disorder Among Oral Medicine And Oral Pathology Specialists
R. Amritha Sripoo1, T. N. Uma Maheswari2*
1 Post Graduate Resident, Department of Oral Medicine and Radiology, SIMATS, Chennai, 600007, India.
2 Professor and Head of the Department of Oral Medicine and Radiology, SIMATS, Chennai, 600007, India.
*Corresponding Author
Dr. T. N. Uma Maheswari,
Professor and Head of the Department of Oral Medicine and Radiology, SIMATS, Chennai, 600007, India.
E-mail: umamaheswaritn@saveetha.com
Received: April 25, 2021; Accepted: October 18, 2021; Published: October 25, 2021
Citation:R. Amritha Sripoo, T. N. Uma Maheswari. Knowledge, Attitude And Perception On Genomic Markers In Oral Potentially Malignant Disorder Among Oral Medicine And Oral Pathology Specialists. Int J Dentistry Oral Sci. 2021;8(10):4842-4847. doi: dx.doi.org/10.19070/2377-8075-21000979
Copyright: T. N. Uma Maheswari©2021. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
Background: A genomic biomarker is a measurement of the expression, function and regulation of a gene that consists of
one or more deoxyribonucleic acid (DNA) and/or ribonucleic acid(RNA) characteristics. Oral potentially malignant disorders
are chronic conditions, which have a higher risk of transformation into oral squamous cell carcinoma.Over years, various
specific and non-specific markers have been introduced that could predict the malignant transformation of Oral Potentially
Malignant Disorders at early stage. Further,early detection plays a crucial role in prognosis and treatment planning.
Aim: This study aims to evaluate the knowledge, attitude, and perception of genomic markers among oral medicine and oral
pathology specialists.
Materials and Method: A 12 multiple choice self constructed questionnaire was prepared by the principal investigator and
the guide circulated among peers for validation. An online survey was created using google forms and distributed among Oral
Medicine and Oral Pathology Specialists. Data was tabulated in Excel spreadsheets. Imported to IBM SPSS software version
2.0 and statistical analysis were performed. Frequency, percentage and chi square test was performed.
Result: On analysis, it was discovered that Oral medicine and Oral pathology specialists were mostly aware about genomic
biomarkers. 42.9% revealed microRNA 21 as the most sensitive microRNA in OPMD, 49.2% revealed easy and convenient
method for Genomic biomarker research in OPMD as Saliva and 43% revealed the major difficulty faced during Genomic
biomarker research in OPMD is Processing of Samples.Majority of the participants believe that genomic biomarkers can
serve as a diagnostic & prognostic tool. of the participants.
Conclusion: Oral medicine and Oral pathology specialists have adequate awareness and knowledge about salivary diagnostic
markers, but certain knowledge has to be brushed up among them. Majority of participants showed a positive attitude towards
its use as a diagnostic & prognostic tool. Furthermore, they need to be trained on these grounds to help them treat their patients
in the best possible way.
Clinical Significance: There is an impervious need for faster ways to detect different pathologies, seeing that many types of
OPMD turning into cancer are discovered only in late stage.miRNAs as a genomic biomarkers can fulfill this, thus being an
impressive research field. Further,the identification of biomarkers provides a novel insight into understanding the mechanisms
of the tumor formation and progression and promising therapy for different cancers.
2.Introduction
3.Materials and Methods
3.Results
4.Discussion
5.Conclusion
5.References
Keywords
Knowledge; Attitude; Perception; Biomarkers; Oral Potentially Malignant Disorders; Oral Pathology.
Introduction
A genomic biomarker is a measurement of the expression, function
and regulation of a gene. A genomic biomarker can consist
of one or more deoxyribonucleic acid (DNA) and/or ribonucleic
acid (RNA) characteristics. Genomic biomarker includes miRNA,
RNA, Mt.DNA, DNA[1, 2]. In recent years miRNAs have
emerged as an important molecules in the complex networks of
gene regulation [3]. These naturally occurring small non coding
RNA molecules that regulate the expression of protein coding
genes at post-transcriptional level have been implicated in a variety
of human disorders, such as infectious diseases, metabolic
disease and malignancy [4]. Several hundred genes in our genome
encode small functional RNA molecules collectively called miRNAs
and are found in normal tissues, blood, and saliva.
miRNA playsan important role in cellular growth, differentiation,
apoptosis, and immune response [5]. During development of
malignancy, some miRNAs are upregulatedand some are downregulated,
so any change in the expression of miRNAs can cause
tumor suppression. Apart from functioning as tumor suppressors,
miRNAs can also promote tumor development (oncogenes) depending
on the tumor types and their specific target protein [5, 6].
The main role of microRNA in the human body is gene regulation.
Given the low survival rate of multiple cancer types, credible
biomarkers for cancer prognosis are urgently required [7].
In recent years, cancer has become the primary cause of mortality
in most countries and regions, and the incidence of human malignancies
has increased substantially [8]. Oral cancer is a preventable
disease as it is mostly preceded by a group of precursor lesions
called oral potentially malignant disorders (OPMDs) [9, 10].
Majority (up to 62%) develop from a visible or invisible OPMD
that is characterized by an increased risk for malignant transformation
(MT) to OSCC [11]. According to the WHO, India is the
second largest consumer and third largest producer of tobacco,
increasing the incidence of OPMD in India which demands the
researchers to involve in studies to find diagnostic biomarkers in
detecting early malignant changes in OPMD.
Scully et al. proved clinical and histopathological assessment of
OPMD is not sufficient to predict malignant transformation,
hence, assessing the miRNA in these lesions would be helpful(
9,12) . Cervigne et al. stated tissue expression of miRNA 21,
miRNA 181b, and miRNA 345 is an early event in leukoplakia
transforming into malignancy [9, 10, 12]. During development of
malignancy, some miRNAs are upregulated and some are downregulated,
so any change in the expression of miRNAs can cause
tumor suppression or act as carcinogens [4]. Each miRNA can
regulate expression of many target genes (multiple proteins) and
expression of each target gene (specific protein) may also be regulated
by multiple miRNAs [13]. miRNA was found to be deregulated
in systemic diseases such as diabetes [14, 15] and hypertension
[16] and was later found to be deregulated in ovarian [17],
breast [18], colon [19], liver [19, 20], and pancreatic cancer tissues
[21]. During the process of development of oral cancer, certain
genes acquire roles in tumorigenesis while some are tumor suppressors.
Increased levels of certain miRNAs cause progression
of malignancy while some suppress malignancy. The upregulated
salivary miRNA 184, and miRNA 21 and downregulated salivary
miRNA 145 can be used as potential biomarkers to predict malignancy.
Of these three, salivary miRNA 184 had the highest sensitivity
and specificity [3]. The aim of this survey was to assess
the knowledge, attitude and perception of genomic biomarkers
among Oral medicine and Oral pathology specialists.
Materials and Methods
This is a cross-sectional study based on self-reported questionnaires.
An online questionnaire was developed by using google
forms, with a consent form appended to it. Link of the questionnaire
was shared through emails, whatsapp and other social
media. Participants with access to the internet could participate in
the study. Participants able to understand english language, who
are pursuing or completed their master degree in Oral Medicine
and Oral pathology and willing to give informed consent were included.
Ethical approval for conducting the survey was obtained
from the Saveetha Dental College and Hospitals. All the data were
collected and compiled by the author. Simple random sampling
was used to select the participants for the study. This provides
equal odds for every member of the population to be chosen as
a participant in the study. The primary items were reviewed by
peers who provided feedback and suggested necessary changes
in order to establish both face and content validity of the survey
questionnaire.
Online self-reported questionnaires developed, which contained
12 multiple choice questions. They were asked to fill in the questionnaire
individually. Responses were. Excel spreadsheets were
used for data collection and manipulation. Age, gender, profession,
knowledge, attitude and perception were assessed. Output
was collected as nominal values, so percentage was calculated and
the results were tabulated. Results were represented by pie charts
and bar graphs.
Data were analyzed using IBM SPSS software version 22. Data
were described using frequencies and percentages. Chi-square was
used to analyze differences between categorical variables. A pvalue
of less than 0.05 was considered statistically significant.
Results
Study was conducted among 35 Oral Medicine specialists and
Oral pathology specialists, out of which 12 were males (34%) and
23 (66%) were female (Table 1) and 17 were Oral medicine specialist(
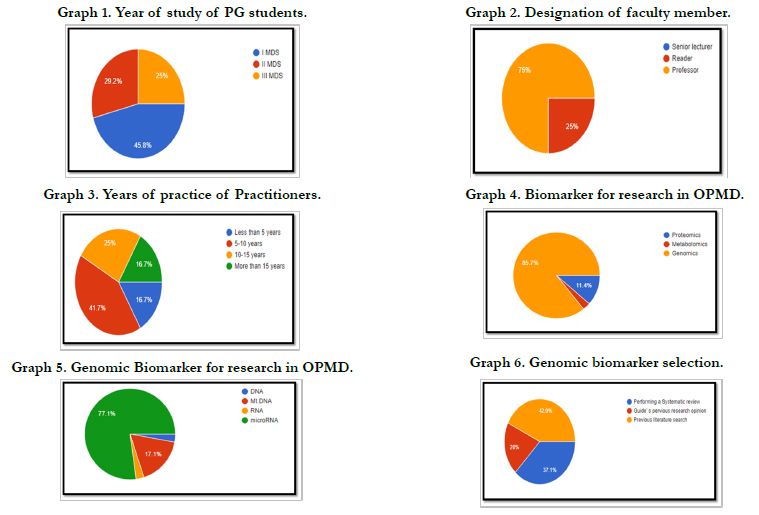
49%) and 18 were oral pathology specialist (51%)(Table 2) 46% were first year MDS students, 29% were second year MDS
students and 25% were third year MDS students, given in Graph
1. 25% were readers and 75% were professors, given in Graph 2.
Among the oral medicine and oral pathology specialists, 17% had
less than 5 years of practice, 42% had 5-10 years of practice, 25%
had 10-15 years of practice and 17% had more than 15 years of
clinical practice; given in Graph 3.
Graph 4 shows participants attitude on biomarker chosen for their
research in OPMD.30 people (86%) chose genomics for their research,
4(11.4%) on proteomics and 1(2.9%) on metabolomics.
Graph 5 shows genomic biomarker chosen for their research in
OPMD. 27 people (77.1%) has selected microRNA for their research,
6(17.1%) selected Mt.DNA, 1(2.9%) selected DNA and
1(2.9%) selected RNA.
Graph 6 shows how genomic biomarker selection was done for
genomic biomarker research in OPMD. 13 people (37.1%) selected
based on performing a Systematic review, 7(20%) selected
based on Guideís previous research opinion and 15(42.9%) selected
based on previous literature research.
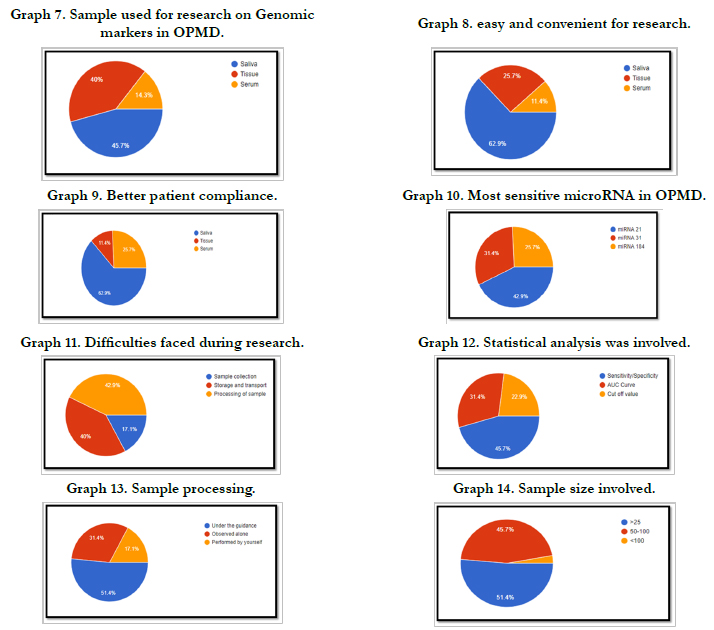
Graph 7 shows Samples used for genomic biomarker research in
OPMD. 16 people (45.7%) used Saliva, 14(40%) used tissue and
5(14.3%) used Serum.
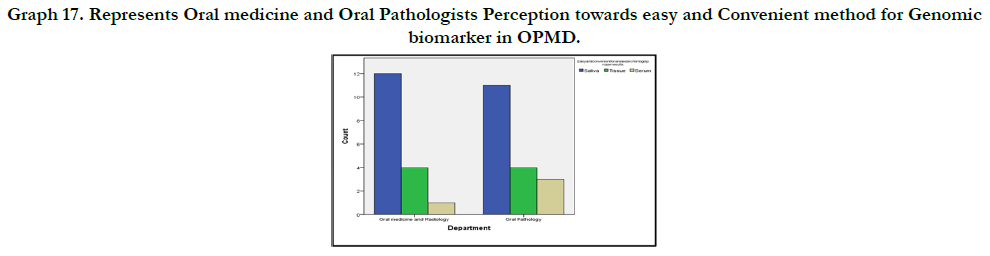
Graph 8 shows easy and convenient one for genomic biomarker
research in OPMD. 22 people (62.9%) suggested Saliva ,9(25.7%)
suggested tissue and 4(11.4%) suggested Serum.
Graph 9 shows samples with better patient compliance for
genomic biomarker research in OPMD. 22 people (62.9%) suggested
Saliva, 9 (25.7%) suggested serum and 4(11.4%) suggested
tissue.
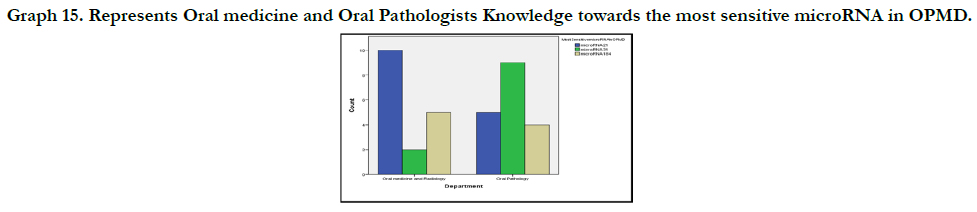
Graph 10 shows the participants knowledge on the most sensitive
biomarker. 15 people (42.9%) suggested miRNA 21, 9(25.7%)
suggested miRNA 184 and 11(31.4%) suggested miRNA 31.
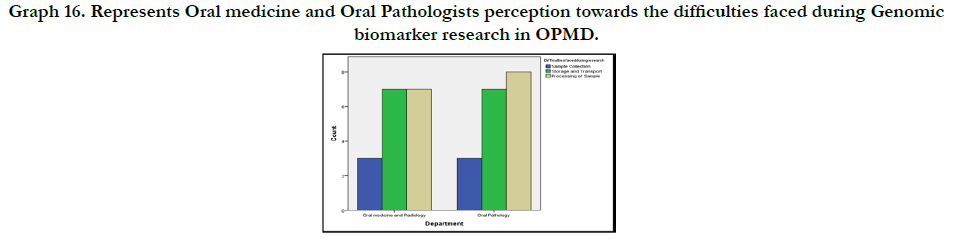
Graph 11 shows the participants perception on difficulties faced
during genomic biomarker research in OPMD. 6 people (17.1%)
suggested Sample collection, 14(40%) suggested Storage and
Transport and 15(42.9%) suggested processing of Sample.
Graph 12 shows statistical analysis involved in genomic biomarker
research in OPMD. 6 (45.7%) Sensitivity/Specificity, 11(31.4%)
AUC Curve and 8(22.9%) Cut off Value.
Graph 12 shows sample processing involved in genomic biomarker
research in OPMD. 18(51.4%) done under the guidance,
11(31.4%) observed alone and 6(17.1%) performed.
Graph 14 shows sample size involved in genomic biomarker
research in OPMD. 18(51.4%) included a sample size of >25,
16(45.7%) included a sample size of 50-100 and 1(2.9%) included
a sample size of <100.
42.9% revealed microRNA 21 as the most sensitive microRNA in
OPMD(Graph 15), 49.2% revealed easy and convenient method
for Genomic biomarker research in OPMD as Saliva (Graph 16)
and 43% revealed the major difficulty faced during Genomic biomarker
research in OPMD is Processing of Samples (Graph 17).
Graph 15. Represents Oral medicine and Oral Pathologists Knowledge towards the most sensitive microRNA in OPMD.
Graph 16. Represents Oral medicine and Oral Pathologists perception towards the difficulties faced during Genomic biomarker research in OPMD.
Graph 17. Represents Oral medicine and Oral Pathologists Perception towards easy and Convenient method for Genomic biomarker in OPMD.
Discussion
This study was conducted among 35 Oral Medicine and Oral Pathology
specialists who has done genomic biomarker research,
out of which 12 were males (34%) and 23 (66%) were female
and 17 were Oral medicine specialist (49%) and 18 were oral
pathology specialist (51%). The results of our study showed that
most of the participants had sufficient knowledge about genomic
biomarkers and difficulties faced during genomic biomarker research
in OPMD. Emerging evidence demonstrates an important
role of miRNAs in regulating diverse cellular processes including differentiation, proliferation and apoptosis [22]. Literature search
revealed no other studies were conducted on knowledge, awareness
and perception of Genomic biomarkers in OPMD. From the
survey results only small sample size is involved in most studies.
The contribution of miRNAs in various types of cancers differs.
Therefore, it is essential to have a larger sample size to be able
to decide between the healthy or diseased status. Saliva contains
many enzymatic enzymes and hence the pathway identification
is difficult whereas tissue is site specific and gives more accuracy
when compared to saliva samples even though Saliva is easy and
convenient and is patient compliance when compared to tissue.
The results of Zahran et al. study revealed upregulatedmiRNA
184 with an area under the curve (AUC) of 0.86 and miRNA 21
with an AUC of 0.73 and downregulated miRNA 145 with an
AUC of 0.68, which proved that these miRNAs are significant
in detecting early malignancy in OPMD and should be further
analyzed in various populations [3]. There is an impervious need
for faster ways to detect different pathologies, seeing that many
types of cancer are discovered in late stage. miRNAs as biomarkers
can fulfill this, thus being an impressive research field(13).
The most important evaluation criteria for circulating miRNAs
as diagnostic and prognostic biomarkers are high sensitivity and
specificity, to avoid false positive or negative diagnosis. An appropriate
biomarker for a specific cancer type should be both
significantly differentially expressed and in correlation with the
outcome of patients. Further, the identification of miRNAs and
their target genes provides a novel insight into understanding the
mechanisms of the tumor formation and progression, and promising
therapy for different cancers (6). Majority of the participants
believed that genomic biomarkers can serve as a diagnostic and
prognostic tool in OPMD. Most of them had a positive attitude
that it can be translated into routine clinical practice. Early detection
of disease plays a crucial role for treatment planning and
prognosis. The most common difficult faced by the researchers
was processing of the samples. Since it was an online study, not
many people from older age groups participated in the survey and
the study was limited to the participants with access to the internet,
who had smartphones and e-mail IDs. And the survey was in
english so people who understand english could only participate.
This survey provides an insight for future researches to fill up the
existing gaps.
Conclusion
Oral medicine and Oral pathology specialists have adequate
awareness and knowledge aboutgenomic biomarkers in Oral
Potentially malignant disorders, but certain knowledge has to be
brushed up among them. Majority of participants showed a positive
attitude towards its use as a diagnostic & prognostic tool. Furthermore, they need to be trained on these grounds to help them
treat their patients in the best possible way. As current techniques
evolve, we anticipate that miRNAs will become a routine approach
in the development of personalized patient profiles, therefore
allowing targeted therapeutic interventions.
References
-
[1]. Institute NC, National Cancer Institute. Genomic Biomarker [Internet].
Definitions. 2020.
[2]. Perkel JM. Genomic Biomarker Discovery: Bringing the Genome to Life [Internet]. Science. 2008.
[3]. Maheswari TNU, Venugopal A, Sureshbabu NM, Ramani P. Salivary micro RNA as a potential biomarker in oral potentially malignant disorders: A systematic review. CiJi Yi XueZaZhi. 2018 Apr-Jun;30(2):55-60. PubmedPMID: 29875583.
[4]. Wu W. MicroRNA and Cancer: Methods and Protocols. Humana Press; 2017. 222 p.
[5]. Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012 Jun 1;33(6):1126-33.
[6]. El-Sakka H, Kujan O, Farah CS. Assessing miRNAs profile expression as a risk stratification biomarker in oral potentially malignant disorders: A systematic review. Oral Oncol. 2018 Feb;77:57-82. PubmedPMID: 29362128.
[7]. Zhang J, Xu A, Miao C, Yang J, Gu M, Song N. Prognostic value of Lin28A and Lin28B in various human malignancies: a systematic review and metaanalysis. Cancer Cell Int. 2019 Apr 2;19:79. PubmedPMID: 30976203.
[8]. Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013 Mar 1;132(5):1133-45. PubmedPMID: 22752881.
[9]. Sankaranarayanan R, Ramadas K, Amarasinghe H, Subramanian S, Johnson N. Oral Cancer: Prevention, Early Detection, and Treatment. In: Gelband H, Jha P, Sankaranarayanan R, Horton S, editors. Cancer: Disease Control Priorities, Third Edition (Volume 3). Washington (DC): The International Bank for Reconstruction and Development / The World Bank; 2015 Nov 1. Chapter 5. PubmedPMID: 26913350.
[10]. Cervigne NK, Reis PP, Machado J, Sadikovic B, Bradley G, Galloni NN, et al. Identification of a microRNA signature associated with progression of leukoplakia to oral carcinoma. Hum Mol Genet. 2009 Dec 15;18(24):4818- 29. PubmedPMID: 19776030.
[11]. Elimairi I, Sami A, Yousef B. Oral Cancer and Potentially Malignant Disorders [Internet]. Histopathology - An Update. 2018.
[12]. Scully C. Challenges in predicting which oral mucosal potentially malignant disease will progress to neoplasia. Oral Dis. 2014 Jan;20(1):1-5. PubmedPMID: 24320967.
[13]. Chattopadhyay E, Singh R, Ray A, Roy R, De Sarkar N, Paul RR, et al. Expression deregulation of mir31 and CXCL12 in two types of oral precancers and cancer: importance in progression of precancer and cancer. Sci Rep. 2016 Sep 6;6:32735. PubmedPMID: 27597234.
[14]. McClelland AD, Kantharidis P. MicroRNAs as Biomarkers of Diabetic Nephropathy [Internet]. Biomarkers in Kidney Disease. 2015. p. 1Ė29.
[15]. Chung AC. microRNAs in Diabetic Kidney Disease. Adv Exp Med Biol. 2015;888:253-69. PubmedPMID: 26663187.
[16]. Marques FZ, Charchar FJ. microRNAs in Essential Hypertension and Blood Pressure Regulation. Adv Exp Med Biol. 2015;888:215-35. PubmedPMID: 26663185.
[17]. Orio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007 Sep 15;67(18):8699-707. PubmedPMID: 17875710.
[18]. Yen MC, Shih YC, Hsu YL, Lin ES, Lin YS, Tsai EM, et al. Isolinderalactone enhances the inhibition of SOCS3 on STAT3 activity by decreasing miRGraph 30c in breast cancer. Oncol Rep. 2016 Mar;35(3):1356-64. PubmedPMID: 26707189.
[19]. Mullany LE, Wolff RK, Herrick JS, Buas MF, Slattery ML. SNP Regulation of microRNA Expression and Subsequent Colon Cancer Risk. PLoS One. 2015 Dec 2;10(12):e0143894. PubmedPMID: 26630397.
[20]. Perilli L, Pizzini S, Bisognin A, Mandruzzato S, Biasiolo M, Facciolli A, et al. Human miRNome profiling in colorectal cancer and liver metastasis development. Genom Data. 2014 Jun 27;2:184-8. PubmedPMID: 26484092.
[21]. Lau C, Kim Y, Chia D, Spielmann N, Eibl G, Elashoff D, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013 Sep 13;288(37):26888-97. PubmedPMID: 23880764.
[22]. Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005 Jul 15;122(1):6-7. PubmedPMID: 16009126.