In silico Pharmacological Screening of Nitrogen containing Bisphosphonate Conjugate with Hydroxyapatite Active Constituents in Finding Potent Inhibitors for Bone Related Diseases
Kalyani A1*, Bharath Srinivasan1, Parasuraman Pavadai2
1 Department of Pharmaceutics, Faculty of Pharmacy, M.S Ramaiah University of Applied Sciences, Bangalore, Karnataka, India.
2 Department of Pharmaceutical Chemistry, Faculty of Pharmacy, M.S Ramaiah University of Applied Sciences, Bangalore, Karnataka, India.
*Corresponding Author
Kalyani A,
Research Scholar, Department of Pharmaceutics, Faculty of Pharmacy, M.S Ramaiah University of Applied Sciences, Bangalore, Karnataka, India.
E-mail: kalyani6009@gmail.com
Received: March 10, 2020; Accepted: July 22, 2020; Published: July 28, 2020
Citation: Kalyani A, Bharath Srinivasan, Parasuraman Pavadai. In silico Pharmacological Screening of Nitrogen containing Bisphosphonate Conjugate with Hydroxyapatite Active Constituents in Finding Potent Inhibitors for Bone Related Diseases. Int J Bone Rheumatol Res. 2020;5(1):77-84. doi: dx.doi.org/10.19070/2470-4520-2000016
Copyright: Kalyani A© 2020. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
Osteoporosis is a skeletal disease-causing bone fragility by disturbing micro architecture of the bone leading to osteoclasts
mediated bone loss. Inhibition of the Farnesyl Pyrophosphate Synthase (FPPS) of mevalonate pathway and Receptor Activator
of Nuclear factor kappa-Β ligand (RANKL/OPG) complex by using anti resorptive drugs like Nitrogen containing
Bisphosphonates aid in the effective treatment of osteoporosis.The in-silico docking analysis of newly synthesized Ibandronate–
hydroxyapatite conjugate has shown as a most powerful binary to HMG-COA Reductase, Farnesyl pyrophosphate
synthase (FPPS), Human Geranyl Geranyl pyrophosphate synthase (GPPS)&RANKL/OPG in contrast to pure Ibandronate.
The docking score of Ibandronate hydroxyapatite (IBA-HAP) was found to be -6.12, -6.75, -5.33 and -6.49 as against standard
pure Ibandronate of -1.28, -2.07, -1.97 and -3.23 kcal/mol. Also,ΔG binding energy and pIC50 values showed promising
potential anti-osteoporotic effect for Ibandronate-hydroxyapatite conjugate.
2.Introduction
3.Materials and Method
4.Results and Discussion
5.Conclusion
6.Acknowledgments
7.References
Keywords
N-Bisphosphonates; Mevalonate pathway; Ibandronate Hydroxyapatite Conjugate; Anti-Osteoporotic Activity; Molecular Modelling.
Introduction
Osteoporosis (OP) is a skeletal disease which thins bone and
weakens it making porous, fragile and finally leading to bone fracture
easily. Majorly low bone mass is seen deterioration of the
micro architectural of bone leading to pain and disability [4].
The concept behind bone becoming weaker and thinner with aging
and disease is due to FPPS, a key regulatory enzyme in the
mevalonate pathway which is responsible for the bone metastases
and osteoclasts mediated bone loss. Anti-resorptive agents
are used in various clinical conditions like hypercalcemia, Paget’s
disease, cancer treatment apart from metabolic bone diseases
[10]. Bisphosphonates are the analogues of pyrophosphate and
are widely used in osteoclasts mediated bone loss due to induced
long term therapy of glucocorticosteroids and also in post-menopausal
women [5]. A step by step deterioration in bone mass
that effects both sexes and due to menopause in women, where
women begin to extremely lose bone mass [11].
Bisphosphonates (BPs)are regarded as most potent drug, widely
used in patients with osteoporosis on osteoclasts suppression [9].
It inhibits FPPS in mevalonate pathway which in turn prevents
osteoclasts by acting on biosynthesis of farnesylation and geranylgeranylation
post translational of signalling proteins where by
inhibiting the activity of osteoclasts and reducing bone resorption
and increasing bone turn over and increase in net bone mass [6].
These BP's are having high binding affinity towards hydroxyapatite
indicating high potency and a long duration of action. Nearly
70% in weight, 50% in volume of human bone contains hydroxyapatite,
commonly known as bone mineral. It contains excellent
biocompatibility and bioactivity with stoichiometric Ca/P
ratio of 1.67 with a hexagonal structure. It is having unique properties
like Osteo conductivity, biocompatibility, bioactivity, nontoxicity
and non-inflammatory nature [12].
BPs, are the bone mineralization endogenous regulators and were
alternatively binding to bone sites for active osteoclasts bone re-sorption, which held by osteoclasts and sequentially inhibit bone
resorption [13]. Cholesterol synthesis of mevalonate pathway is
a bio synthetic pathway responsible for osteoclasts inactivation
which is crucial in achieving the therapeutic target [1] by inhibitingsome
of the isoprenoid lipids like HMG CO-A synthase,
HMG CO-A reductase, mevalonate kinase, Human Geranyl Geranyl
pyrophosphate synthase which bound to Geranyl Geranyl
pyrophosphate (GGPP) that areessential for the activation of
prenylation of small GTPase isoprenoids to prenylation GTPase
signalling protein that helps in normal functioning of proteins [7].
Another protein target Cytokine receptor triad Human RANKL/
OPG a bone morphogenic protein (BMP-2) that crucially controls
the terminal differentiation step of osteoblasts and osteoclasts
also binds to a protein secreted by the cells of osteoblasts
linage known as Osteoprotegerin [2].
In the present paper, the comparative docking analysis of newly
synthesized Ibandronate–hydroxyapatite conjugate and pure
Ibandronate (N-Bisphosphonate drug) ligand with target proteins
like mevalonate pathway enzymes and RANKL/OPG have been
discussed.
Materials and Method
Autodock works on Monte Carlo simulated annealing and
Lamarckian genetic algorithm (LGA) to create a set of probable
conformations. LGA method was used as a global optimizer and
energy minimizer. The docking studies were carried out by using
Autodock 4.2 suite to predict binding of molecules to the target
protein structure [8]. Apart from generating the binding energies
of the docked molecules, nature and the position of ligand in the
binding site can be visualized. In this docking simulation, semiflexible
docking protocols have been used in which the target
proteins were set aside as rigid. The drug substrate to be docked were kept flexible, in order to discover random number of torsional
degrees of freedom in addition to the six spatial degrees of
freedom spanned by the translational and rotational parameters.
The ligand molecular structure of Ibandronate and Ibandronatehydroxyapatite
conjugate were drawn using chem draw Ultra
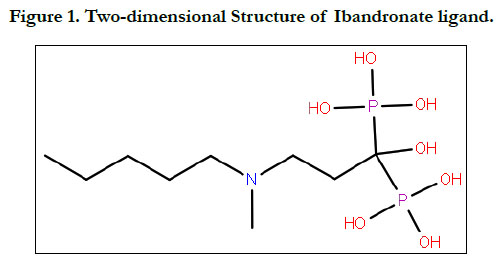
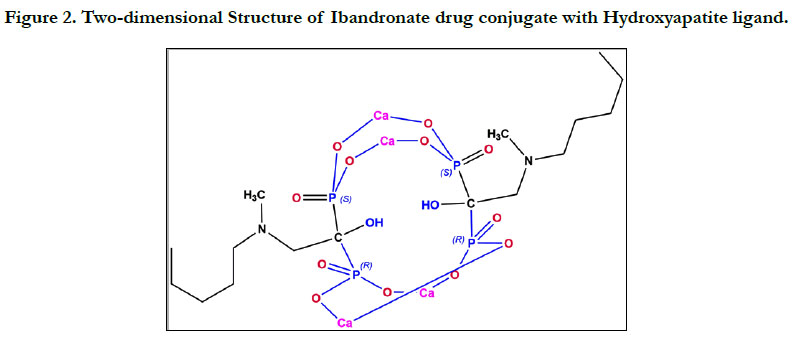
7.0.4. (Figure 1 & 2).
The crystal structure of protein receptors with PBD ID’s – 3SQZ
(HMG_CoA synthase), 1DQ9 (Human HMG-COA Reductase),
2F7M (Human Farnesyl Pyrophosphate synthase (FPPS))[13],
2HFU (Mevalonate kinase), 2Q80 (Human Geranyl Geranyl pyrophosphate
synthase bound to GGPP), of Mevalonate pathway
and 3URF (Human RANKL/OPG complex), solved using X- ray
diffraction method obtained from RSCB protein data bank (PDB)
(http://www.rcsb.org/). Protein structure were subjected for protein
preparation using BOVIA Discovery Studio 2017 R2 Client
software. He-atoms and water molecules were removed from the
protein structure and subjected to applied force field in order to
minimize the energy of the atoms present in the structure and
made them stable to react with the ligands. Hydrogen atoms were
added, Gasteiger chargers were assigned by merging non polar
bonds, followed by assigning of AD4 type to the protein receptor.
Ligands were defined for rigid roots of Ibandronate and Ibandronate
conjugate hydroxyapatite automatically. The grid box size
was set at 90, 90, and 90 A° (X, Y, and Z), depending on the protein
size. The Lamarckian Genetic Algorithm (LGA) was chosen
to search for the best conformers. The size of the population was set to 100 and the individuals were initialized arbitrarily. Energy
estimation was set to 500000, maximum number of generations
1000, maximum number of top individual that automatically survived
was set to 1to 10 LGA runs were performed [3].
Receptor grid was generated to determine the size of the active
site. The most probable orientation of the ligands in the binding
pocket is identified and a scoring function is used to quantify the
strength ofthe interaction in a molecule can make in a particular
orientation. In this the grid box is prepared by taking X, Y,
Z dimensions as 90,90,90 respectively centred the grid box on
macromolecule. The precision was favoured over the standard
mode sequentially give a better correlation between good poses
and good scores.
Results and Discussion
The docking analysis was done for the ligands with the target
proteins RANKL/OPG, Mevalonate pathway enzymes using the
docking software Autodock and the docked images are shown.
The structures dockedby Autodock are generally ranked according
to the Binding energy function & pIC50 values. Autodock program
scoring function is presented in the kcal/mol form. The
method of evaluating the accuracy of a docking procedure is todetermine
how closely the lowest energy pose (binding conformation)
predicted by the object scoring function.
To study the binding interaction and binding affinity of the Ibandronate
and Ibandronate hydroxyapatite conjugate to HMG-CoA
synthase, Human HMG-COA Reductase, FPPS, Mevalonate kinase,
GGPS of Mevalonate pathway and Human RANKL/OPG
complex, both the ligands were docked into theactive site ofproteins.
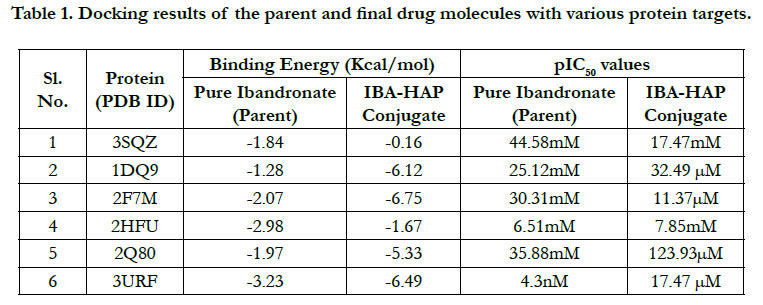
The docking result of these ligands is given in Tables 1 and
2. The interaction energywere calculated for each complex.
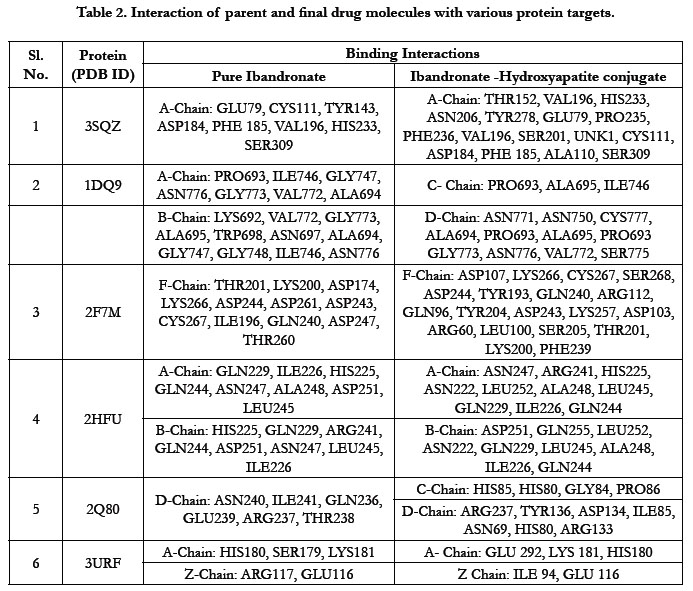
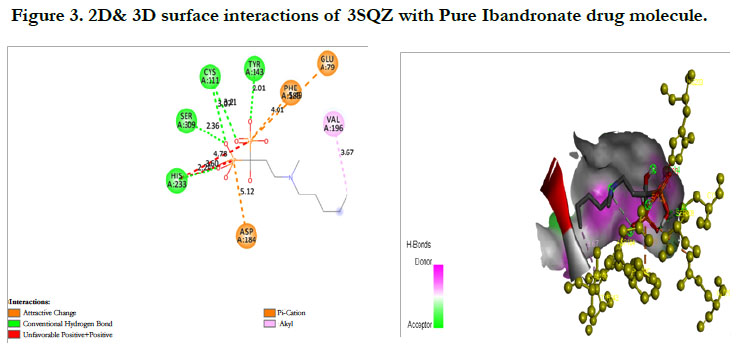
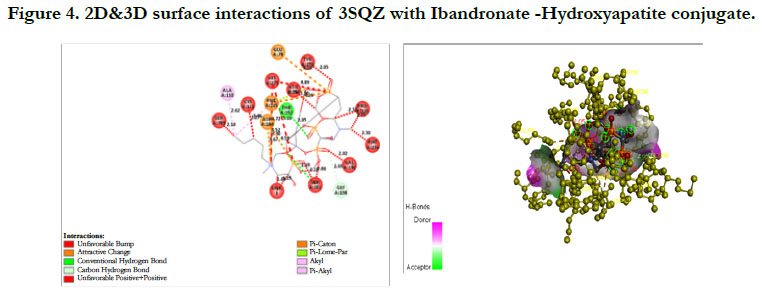
The docking score by using Autodock has shown -1.84 kcal/ mol, -0.16 kcal/mol for Ibandronate ligand and Ibandronate hydroxyapatite conjugate ligand with HMG COA synthase(PDB ID: 3SQZ) shown the phosphate groups of the parent Ibandronate molecule had formed conventional hydrogen bonding with HIS233, SER309, CYS111, TYR143 of A-chain amino acid residues; the alkyl group had formed alkyl bonding with VAL196 of A chain amino acid residues; the oxygen atom of phosphate group had undergone attractive charges with PHE185, GLU79, ASP184 of A-chain amino acid residues (Figure 3), whereas the interactions of Ibandronate hydroxyapatite conjugate molecule with the protein receptor of HMG co A synthase shown, the oxygen atoms of phosphate groups had formed an unfavourable positive-positive interactions with SER309, CYS111, PHE185, HIS233, ASN206, TYR278, PRO235, PHE236, VAL196, SER201; the phosphate atom of the molecule had undergone attractive charges and pi-cation interactions with GLU79, PHE185, ASP184 and a conventional hydrogen bonding was found with another oxygen atom of the molecule with THR152 of A-Chain amino acid residues (Figure 4).
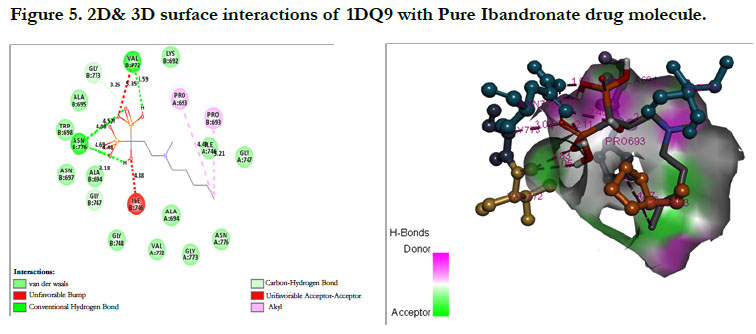
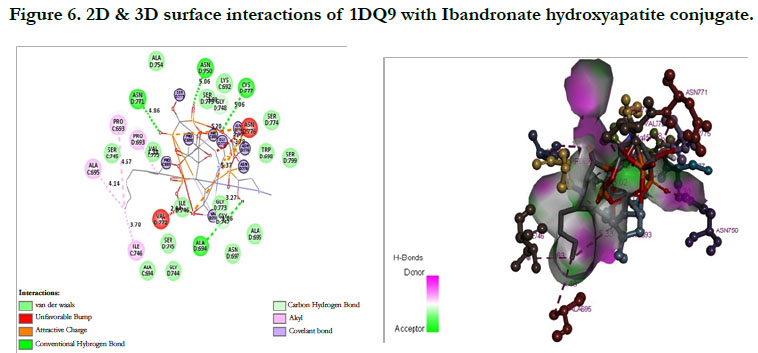
Ibandronate ligand and Ibandronate hydroxyapatite conjugate ligand with Human HMG-COA Reductase (PDB ID: 1DQ9) shown docking score -1.28 kcal/mol & -6.12 kcal/mol, the phosphate groups of the parent molecule Ibandronate had formed conventional hydrogen bonding with VAL772, ASN776 of Bchain amino acid residues; the alkyl group had formed alkyl bonding with PRO693 of A chain and PRO693 of B-chain amino acid residues; the oxygen atom of phosphate group had undergone carbon hydrogen bonding with GLY773 of B-chain amino acid residues (Figure 5), the interactions of conjugate molecule with the protein receptor shown the oxygen atoms of phosphate groups had undergone conventional hydrogen bonding with ASN771, ASN750, CYS777, ALA694 of D-Chain amino acid residues; the alkyl side chain had undergone alkyl bonding with PRO693 of D-chain amino acid residue and PRO693, ALA695, ILE746 of C- Chain amino acid residues and covalent bonding with PRO693 of C- Chain amino acid residue and GLY773, ASN776, VAL772, SER775 OF D-Chain amino acid residues (Figure 6).
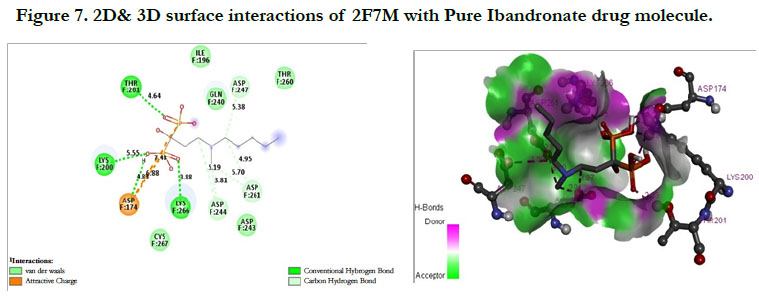
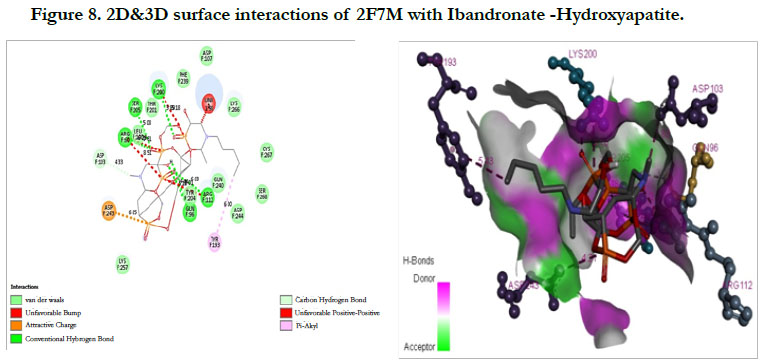
The docking score shown -2.07 kcal/mol, -6.75 kcal/mol for Ibandronate ligand and Ibandronate hydroxyapatite conjugate ligand with Human Farnesyl Pyrophosphate synthase (FPPS) (PDB ID: 2F7M) shows, the phosphate groups of the parent molecule had formed conventional hydrogen bonding with THR201, LYS200, LYS266 of F-chain amino acid residues; the alkyl chain of parent molecule had formed carbon hydrogen bonding with ASP247, ASP261, ASP244 of F-chain amino acid residues; the phosphorus atom of phosphate group had undergone attractive charges with ASP174 of F-chain amino acid (Figure 7) the interactions of conjugate with the protein receptor shown, the alkyl side chain had undergone pi-alkyl interactions with TYR193 of F-chain amino residue; the oxygen atoms of phosphate groups had undergone conventional hydrogen bonding with ARG 112, GLN96, ARG60, SER205, LYS200 of F-chain amino acid residues; the dihydro pyridine of conjugate had undergone attractive charge interaction with ASP243 of F-chain amino acid residue (Figure 8).
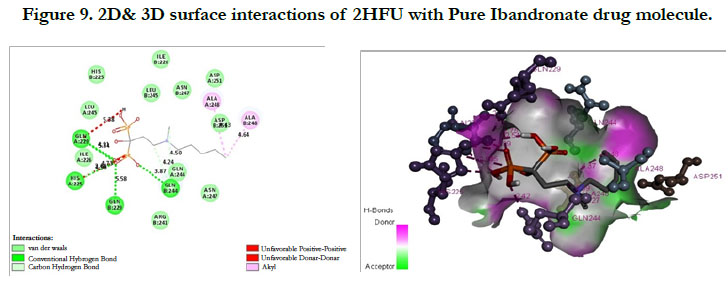
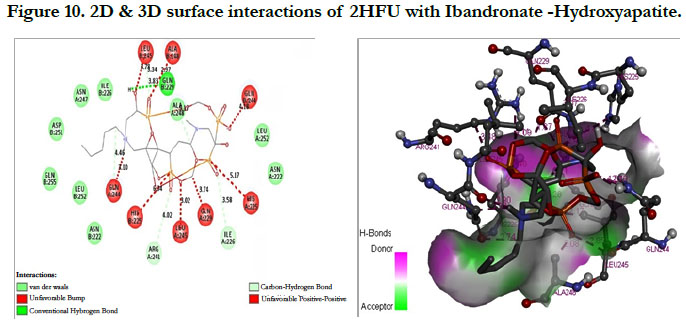
The docking score shown -2.98 kcal/mol, -1.67 kcal/mol for Ibandronate ligand and Ibandronate hydroxyapatite conjugate ligand with protein receptor Mevalonate kinase (PDB ID: 2HFU) shows the phosphate groups of the Ibandronate molecule had formed conventional hydrogen bonding with GLN229, HIS225 of A-chain amino acid residues and GLN229, GLN244 of Bchain amino acid residues; the alkyl chain of parent molecule had formed alkyl bonding with ALA248 of A chain and ALA248 of B-chain amino acids (Figure 9) The interactions of Ibandronate hydroxyapatite conjugate shows, the oxygen atoms of phosphate groups had undergone unfavourable positive-positive interactions with GLN244, ALA248, LEU245, GLN244, HIS225, GLN229, HIS225, LEU245 of A&B-chain amino acid residues respectively; another oxygen atom of phosphate group had undergone carbon hydrogen bonding with ILE226, ARG241 of A-chain amino acid residue; the hydroxyl group had undergone conventional hydrogen bonding with GLN229 of B-chain amino acid residue (Figure 10).
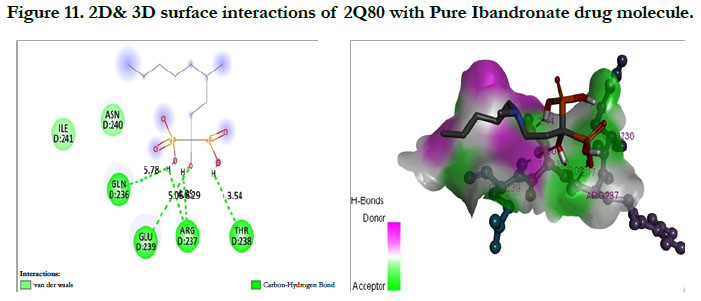
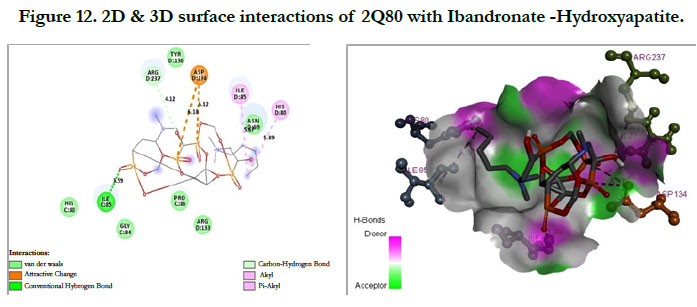
The interactions of Ibandronate molecule with the protein receptor with PDB ID: 2Q80 (Human Geranyl Geranyl pyrophosphate synthase bound to GGPP), shows the oxygen atom of phosphate groups of the Ibandronate molecule had formed conventional hydrogen bonding with GLN236, GLU239, ARG237, THR238 amino acids (Figure 11) with the docking score -1.97 kcal/mol and -5.33 kcal/mol for the Ibandronate Hydroxyapatite conjugate and the interactions are, the oxygen atoms of phosphate groups had undergone conventional hydrogen bonding with ILE85 of C-chain amino acid residues; another oxygen atom of phosphate group of conjugate had undergone carbon hydrogen bonding with ARG237 of D-chain amino acid residue; the phosphate group of final molecule had undergone attractive charge interaction with ASP134 of D-chain amino acid residue; the alkyl side chain had undergone alkyl and pi-alkyl interactions with HIS80, ILE85 of D-chain amino acid residues (Figure 12).
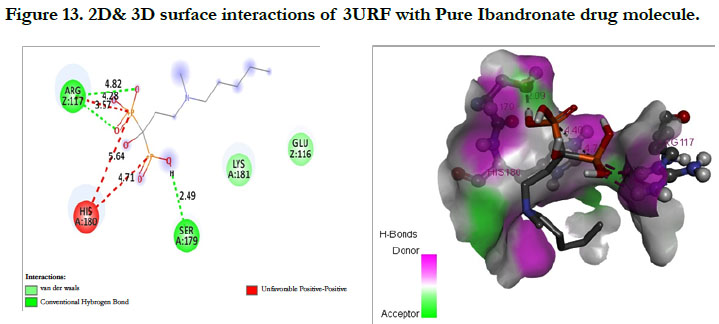
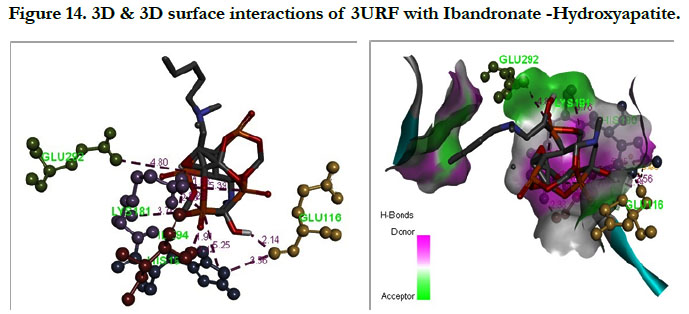
The docking score shown -3.23 kcal/mol, -6.49 kcal/mol for Ibandronate ligand and Ibandronate hydroxyapatite conjugate ligand with protein receptor 3URF. The interactions of Ibandronate molecule with the protein receptor with Human RANKL/OPG complex shown, the oxygen atom of phosphate groups of the parent molecule had formed conventional hydrogen bonding with SER179, ARG117 of A&Z-chain amino acid residues respectively; another oxygen atom of phosphate group had undergone unfavourable positive-positive interactions with HIS180 of A-chain amino acid residues (Figure 13). The interactions of Ibandronate hydroxyapatite conjugate with the protein receptor with PDB ID: 3URF are, the Ibandronate hydroxyapatite conjugate had undergone bonding with GLU292, LYS181, HIS180 of A -chain amino acid residues and ILE94, GLU116 ofZ-chain amino acid residues respectively. The 3D interactions and 3D surface interactions (Figure 14).
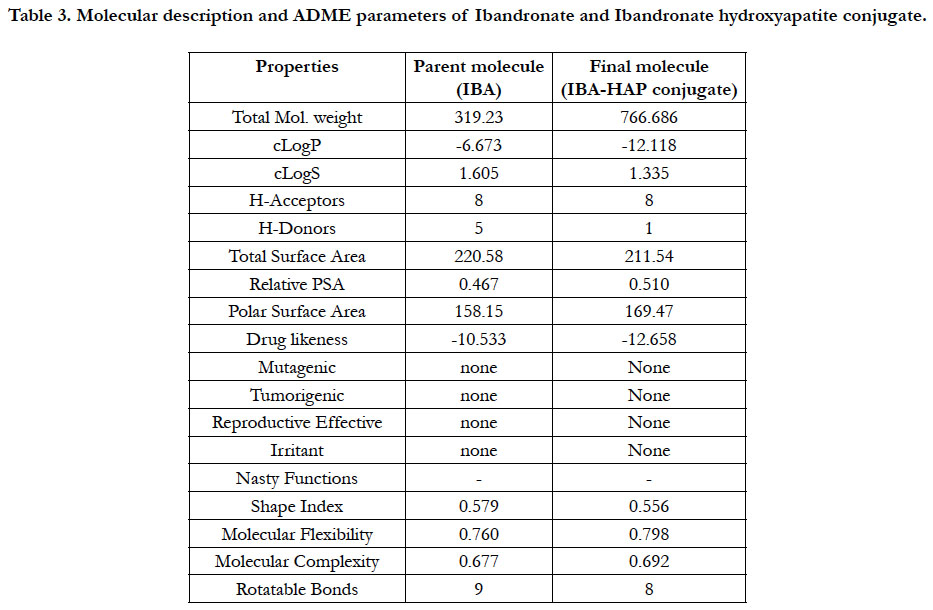
Docking simulation at the active sites with proteins 3SQZ, 1DQ9, 2F7M, 2HFU, 2Q80, 3URF with respect to pure Ibandronate & Ibandronate-hydroxyapatite conjugate docking simulation score and pIC50 values were recorded in Table 1. Docking studies performed by Autodock 4.2 was confirmed that above inhibitors fit into the binding pocket of the HMG_CoA synthase, Human HMG-COA Reductase, FPPS, Mevalonate kinase, GPPS of Mevalonate pathway and Human RANKL/OPG complex, receptors are shown in the Figures: 4-15. We observe successful docking,intermolecular hydrogen bonding and lipophilic interactions betweenthe ligand and receptor. Mostly the oxygen atoms of phosphate groups had undergone conventional hydrogen bonding with Amino acid residues of targets. On the basis of minimum energy concept, it can be concluded that ΔG binding energy and pIC50 values shows promising anti-osteoporotic effect for Ibandronate-Hydroxyapatite conjugate and it can act as a most powerful to inhibit FPPS, GPPS & RANKL/OPG in contrast to pure Ibandronate. Data warrior the drug- conjugate also complied with Lipinski’s rule without any compliances for mutagenicity, irritability and tumorigenicity (Table 3).
Table 3. Molecular description and ADME parameters of Ibandronate and Ibandronate hydroxyapatite conjugate.
Conclusion
The newly synthesized drug molecule IBA-HAP has shown high
binding and powerful inhibitory activity towards Mevalonate
pathway enzymes like Human HMG-COA Reductase, FPPS,
GGPS and HUMAN RANKL/OPG proteins as compared to
Pure Ibandronate. Based on the in-silico studies, the results support
in designing of new drugs to treat osteoporosis in which
potential inhibitors will interact strongly with residues.
Acknowledgments
The authors are thankful to the management of M.S. Ramaiah
University of Applied Sciences for providing required support to
carry out the research work.
References
- Amin D, Cornell SA, Perrone MH, Bilder GE. 1-Hydroxy- 3-(methylpentylamino)-propylidene-1,1-bisphosphonic acid as a potent inhibitor of squalene synthase. Arzneimittelf orschung. 1996;46(8):759-762. Pubmed PMID:9125274.
- Çankaya M, Cizmeci Şenel F, Kadioglu Duman M, Muci E, Dayisoylu EH, Balaban F. The effects of chronic zoledronate usage on the jaw and long bones evaluated using RANKL and osteoprotegerin levels in an animal model. Int J Oral Maxillofac Surg. 2013;42(9):1134-1139. Pubmed PMID:23522850.
- Jagyasi A, Choubey J, Patel A, Verma MK. Molecular Modeling and Docking Analysis of Novel Drug like Compounds for NDM-1. Int J Chem Anal Sci. 2013;1:47-54.
- Dang L, Liu J, Li F, Wang L, Li D, Guo B, et al. Targeted delivery systems for molecular therapy in skeletal disorders. Int J Mol Sci.2016 Mar;17(3):428. Pubmed PMID:27011176.
- Dunford JE, Thompson K, Coxon FP, Luckman SP, Hahn FM, Poulter CD,, et al. Structure-activity relationships for inhibition of farnesyl diphosphate synthase in vitro and inhibition of bone resorption in vivo by nitrogencontaining bisphosphonates. J Pharmacol Exp Ther. 2001;296(2):235-242. Pubmed PMID:11160603.
- Frith JC, Mönkkönen J, Auriola S, Mönkkönen H, Rogers MJ. The molecular mechanism of action of the antiresorptive and antiinflammatory drug clodronate: evidence for the formation in vivo of a metabolite that inhibits bone resorption and causes osteoclast and macrophage apoptosis. Arthritis Rheum. 2001;44(9):2201-2210.Pubmed PMID:11592386.
- Luckman SP, Hughes DE, Coxon FP, Graham R, Russell G, Rogers MJ. Nitrogen-containing bisphosphonates inhibit the mevalonate pathway and prevent post-translational prenylation of GTP-binding proteins, including Ras. J Bone Miner Res. 1998;13(4):581-589.Pubmed PMID:9556058.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of computational chemistry. 1998 Nov 15;19(14):1639-62.
- Nancollas GH, Tang R, Phipps RJ, Henneman Z, Gulde S, Wu W,,et al. Novel insights into actions of bisphosphonates on bone: differences in interactions with hydroxyapatite. Bone. 2006;38(5):617-627.Pubmed PMID:16046206.
- Rodan GA, Fleisch HA. Bisphoshonates:Mechanism of action.J Clin Invest. 1996; 97:2692-2693.
- Rodan GA, Martin TJ. Therapeutic approaches to bone diseases. Science. 2000 Sep 1;289(5484):1508-14.
- Russell RG, Watts NB, Ebetino FH, Rogers MJ. Mechanisms of action of bisphosphonates: similarities and differences and their potential influence on clinical efficacy. Osteoporos Int. 2008;19(6):733-759.Pubmed PMID:18214569.
- Widler L, Jaeggi KA, Glatt M, Müller K, Bachmann R, Bisping M, et al. Highly potent geminal bisphosphonates. From pamidronate disodium (Aredia) to zoledronic acid (Zometa). J Med Chem. 2002;45(17):3721-3738. Pubmed PMID:12166945.